More Information
Submitted: January 01, 2021 | Approved: January 25, 2021 | Published: January 27, 2021
How to cite this article: Nikhra V. Exploring COVID-19: Relating the spike protein to infectivity, pathogenicity and Immunogenicity. Int J Clin Virol. 2021; 5: 001-010.
DOI: 10.29328/journal.ijcv.1001029
ORCiD: orcid.org/0000-0003-0859-5232
Copyright License: © 2021 Nikhra V. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: ACE2 receptors; COVID-19; Neutralising antibodies; Receptor binding domain; SARS-CoV-2; SNVs; Spike protein; SARS-CoV-2 mutants; D614G; B.1.1.7; VUI-202012/01
Exploring COVID-19: Relating the spike protein to infectivity, pathogenicity and Immunogenicity
Vinod Nikhra*
Senior Chief Medical Officer and Consultant, Department of Medicine, Hindu Rao Hospital and NDMC Medical College, New Delhi, India
*Address for Correspondence: Dr. Vinod Nikhra, Senior Chief Medical Officer and Consultant, Department of Medicine, Hindu Rao Hospital and NDMC Medical College, New Delhi, India, Email: [email protected]; [email protected]
Introduction: SARS-CoV-2 life cycle: The disease which reportedly began in Chinese city Wuhan in November-December 2019 manifesting as severe respiratory illness, soon spread to various parts of the world, and was named COVID-19, and declared a pandemic by WHO. The life cycle of SARS-CoV-2 begins with membrane fusion mediated by Spike (S) protein binding to the ACE2 receptors. Following viral entry and release of genome into the host cell cytoplasm there occurs replication and transcription to generate viral structural and non-structural proteins. Finally, VLPs are produced and the mature virions are released from the host cell.
Immunogenicity of the spike protein: The S protein is considered the main antigenic component among structural proteins of SARS-CoV-2 and responsible for inducing the host immune response. The neutralising antibodies (nAbs) targeting the S protein are produced and may confer a protective immunity against the viral infection. Further, the role of the S protein in infectivity also makes it an important tool for diagnostic antigen-based testing and vaccine development. The S-specific antibodies, memory B and circulating TFH cells are consistently elicited following SARS-CoV-2 infection, and COVID-19 vaccine shots in clinical trials.
The emerging SARS-CoV-2 variants: The early genomic variations in SARS-CoV-2 have gone almost unnoticed having lacked an impact on disease transmission or its clinical course. Some of the recently discovered mutations, however, have impact on transmissibility, infectivity, or immune response. One such mutation is the D614G variant, which has increased in prevalence to currently become the dominant variant world-over. Another, relatively new variant, named VUI-202012/01 or B.1.1.7 has acquired 17 genomic alterations and carries the risk of enhanced infectivity. Further, its potential impact on vaccine efficacy is a worrisome issue.
Conclusion: THE UNMET CHALLENGES: COVID-19 as a disease and SARS-CoV-2 as its causative organism, continue to remain an enigma. While we continue to explore the agent factors, disease transmission dynamics, pathogenesis and clinical spectrum of the disease, and therapeutic modalities, the grievous nature of the disease has led to emergency authorizations for COVID-19 vaccines in various countries. Further, the virus may continue to persist and afflict for years to come, as future course of the disease is linked to certain unknown factors like effects of seasonality on virus transmission and unpredictable nature of immune response to the disease.
The disease which reportedly began in Chinese city Wuhan in November-December 2019 manifesting as severe respiratory illness, soon spread to various parts of the world, and was named COVID-19, and declared a pandemic disease by the World Health Organization (WHO). The disease further spread to certain Arabian and European countries. While these countries were recovering from the epidemic, the disease took hold in the UK and USA, and the South American, Arabian, and South Asian countries, predominantly affecting Brazil, Peru, Iran, and India. Presently, most of the European countries are witnessing recurrent outbreaks and a resurgence of COVID-19, whereas the epidemic is not yet over in other countries [1]. This calls for a need for exploring further the factors affecting the disease transmission, pathogenicity, and immunogenicity, apart from rigorous preventive and control measures, and vigorous vigilance and surveillance.
The life cycle of SARS-CoV-2
The life cycle of SARS-CoV-2 begins with membrane fusion mediated by conformational changes in the Spike (S) glycoprotein triggered by ACE2 receptor binding. The cleavage at the S1/S2 site yields a surface subunit S1, which attaches the virus to the host cell ACE2 receptor, and a transmembrane subunit S2, which mediates the fusion of viral and host cell membranes to facilitate viral entry. Following entry, SARS-CoV-2 releases its genomic RNA into the host cell cytoplasm.
Genome RNA is first translated into viral replicase polyproteins (pp1a and 1ab), which are further cleaved by viral proteases into some 16 non-structural proteins. A replication-transcription complex (RTC) is formed based on the non-structural proteins. In the process of genome replication and transcription mediated by RTC, the negative−sense genomic RNA is synthesized and used as a template to produce positive-sense genomic RNA and sub-genomic RNAs.
The nucleocapsid (N) structural protein and viral RNA are replicated, transcribed, and synthesized in the cytoplasm, whereas other viral structural proteins, including the S protein, membrane (M) protein and envelope (E) protein, are transcribed and then translated in the rough endoplasmic reticulum (RER) and transported to the Golgi complex.
In the RER and Golgi complex, the SARS-CoV-2 glycoprotein is subjected to co-translational and post-translational processing, including signal peptide removal, trimerization, extensive glycosylation and subunit cleavage. The N protein is subsequently associated with the positive sense genomic RNA to become a nucleoprotein complex (nucleocapsid). While S, M, and E proteins as well as other viral proteins are further assembled, followed by budding into the lumen of the ER-Golgi intermediate compartment (ERGIC) to form mature virions.
Finally, the mature virions are released from the host cell.
Exploring SARS-CoV-2 S protein
The virus and spike (S) protein: The novel coronavirus, SARS-CoV-2 is a single-stranded RNA-enveloped virus. Its RNA-based genome is 29,881 bp in length and encodes 9860 amino acids. The SARS-CoV-2 S protein, primarily facilitating the viral entry into the host cells, is a 1273 amino acid homo-trimeric class I fusion protein. The third open reading frame (ORF) in the SARS-CoV-2 genome encodes the S protein, which is the largest protein in the group of four structural proteins including M, E and N proteins [2]. The SARS-CoV-2 S protein bears 76% amino acid sequence identity with SARS-CoV and 93% and 97% amino acid identity with that of the Bat-CoV RaTG13 and Pangolin-CoV, respectively [3]. Due to the conserved residues and binding to the receptors, immunity to SARS-CoV appears to confer a limited immunity to SARS-CoV-2 [4].
The SARS-CoV-2 S protein identifies a transmembrane protein, neuropilin-1 on the surface of human cells, through which it facilitates the virus to attach, invade and infect the host cells [5]. The neuropilins are adhesion molecules responsible for cellular activities such as cell adhesion, survival, repulsion, and attraction.
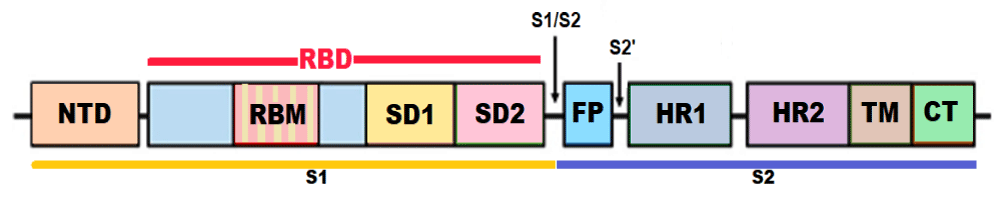
The structure of S protein: The S protein has a size of 180–200 kDa and length of 1273 aa. The S protein trimers visually form a characteristic bulbous, crown-like protrusions surrounding the viral particle. The S protein is composed of various functional regions or domains. The globular head, S1 subunit contains the N-terminal domain (NTD) and receptor-binding domain (RBD), whereas the stalk, S2 subunit contains the C-terminal (CT) membrane fusion domain followed by the two heptad regions (HR1 and HR2), the transmembrane domain (TMD) and the cytosolic tail (CT). The S protein ectodomain is heavily glycosylated with heterogeneous N-linked glycans and exists in a prefusion and a post-fusion conformation. The associated oligosaccharides influence priming by host proteases and conceal it from antibody recognition [6].
The RBD binds to the aminopeptidase N (APN) region of ACE2 receptors in the host cells [7]. APN is transmembrane glycoprotein expressed on the apical membranes of epithelial cells in the respiratory and enteric tracts, endothelial cells, and renal cells; at synaptic junctions in neural tissues; and on cells of the immune system. While the S1 RBD domain is a highly mutable region, the HR region of the S2 subunit is conserved among HCoVs. The mutations of key residues in RBD can enhance the interactions with ACE2 [8]. The S1 subunit binds with ACE2 to promote the formation of endosomes and triggers the viral fusion activity. The S2 subunit, composed successively of a FP, HR1, HR2, TM domain, and cytoplasmic tail domain (CT), is responsible for viral fusion and entry. The HR1 and HR2 are composed of a repetitive heptapeptide: HPPHCPC, where H is a hydrophobic or traditionally bulky residue, P is a polar or hydrophilic residue, and C is a charged residue (Figure 1).
Figure 1: Structure of the SARS-CoV-2 S protein (NTD - N-terminal domain, RBM – Receptor binding motif, SD1 - subdomain 1, SD2 - subdomain 2, FP - fusion peptide, HR1 - heptad repeat 1, HR2 - heptad repeat 2, TM - transmembrane region, and CT – Cytoplasmic tail or IC - intracellular domain).
The S protein exists in a metastable conformation and when the virus interacts with the host cell, extensive structural rearrangement of its components occurs, allowing the virus to fuse with the host cell membrane. The switch in conformations is triggered by RBD-ACE2 receptor binding, which exposes various regions within the S2 subunit [9].
ACE2 receptor binding and viral fusion: To initiate cellular entry, while engaging with ACE2 receptor, the RBD moves like a hinge between two conformations (‘up’ or ‘down’) to expose or hide the residues that bind the APN region. Within the RBD, there is a receptor binding motif (RBM) which makes the primary contact with the carboxypeptidase domain of ACE2. SARS-CoV-2 binds to ACE2 with higher affinity than SARS-CoV owing to the presence of a 4-residue motif in the RBM that makes a better contact with ACE2 than the SARS-CoV S protein. ACE2 as receptors, however, have differential tissue tropism and the structural variations in ACE2 may influence its binding with the S protein [10].
There occurs change in S2 conformation with the RBD bonding with ACE2, which exposes the cleavage sites to cellular proteases, followed by cleavage of the S protein by transmembrane protease serine 2 (TMPRSS2) and other cellular proteases. The cleavage initiates the insertion of FP into the host cell membrane, exposure of the pre-hairpin coiled-coil of the HR1 domain and its interaction with the HR2 to form the six-helical bundle (6-HB). HR1 forms a homo-trimeric assembly in which three highly conserved hydrophobic grooves on the surface binding to HR2 are exposed and bring the viral envelope and cell membrane into proximity for fusion and entry [11].
Whereas other CoVs spike proteins are cleaved at the junction between S1 and S2, the SARS-CoV2 has an additional distinct protease (furin) cleavage site (S2’). With the binding of S1 to the receptor, S2 undergoes an additional cleavage by host proteases through exposure of a second cleavage site (S2′). In the two-step process, the cleavage at the S2′ site activates the protein for membrane fusion and allows fusion of the viral membrane with the host cell membrane. Apart from TMPRSS2, the SARS-CoV-2 S protein can be proteolytically activated by a variety of other cellular proteases including cathepsin B and L (endosomal cysteine proteases), furin, elastase, factor X and trypsin. The protease induced priming and proteolysis initiate the process of cellular entry.
The second cleavage site (S2’) changes the conformation of the S protein from the prefusion to the post-fusion state and expands the versatility of SARS-CoV-2 for cleavage by cellular proteases, the tropism and transmissibility owing to the wide cellular expression of furin proteases, especially in the respiratory tract [12]. This also allows the newly synthesised virions can be secreted in a ‘preactivated’ state ready to fuse with and infect other cells by releasing the FP to disrupt the host cell bilayer-lipid membrane lipid bilayers to prime for fusion without the need to bind to a cellular receptor, such as ACE2. The SARS-CoV-2 S protein has thus evolved further to exploit respiratory cell receptors and proteases to enable enhanced infectivity and rapid spread.
The steps in viral invasion and internalisation, thus, include binding of the virus to the cell surface, alteration of the conformation of the S protein, proteolysis of the S protein, and release of the S2 subunit followed by fusion of the virion and endocytosis. The S protein binds to the host cell by recognizing the receptor ACE2, which is a homolog of ACE and distributed in the lungs, intestines, heart and vasculature, and kidneys. The alveolar epithelial type II cells are the major expressing cells. SARS-CoV-2 S binds to human ACE2 with a dissociation constant (KD) of 14.7 nM, which for SARS-CoV S is 325.8 nM, indicating that SARS-CoV-2 S is more sensitive to ACE2 than is SARS-CoV S [13].
The S subunits exist in a noncovalent form in the uncleaved state and cleavage of S1 and S2 subunits by host proteases, initiates the fusion process. The proteases furin and neuropilin-1 (NRP1), which are abundantly expressed in the respiratory and olfactory epithelium, with highest expression in endothelial and epithelial cells, bind the substrates and potentiate the SARS-CoV-2 infectivity [14]. Trypsin is another host cell protease that can cleave the S protein. Viral fusion process refers to fusion of the viral membrane and host cell membrane, followed by internalisation and the release of the viral genome into the host cell.
Immunogenicity of the s protein
The experimental immunological studies: The S protein is considered the main antigenic component among structural proteins of SARS-CoV-2 and responsible for inducing the host immune response. The neutralising antibodies (nAbs) targeting the S protein are produced and may confer a protective immunity against the viral infection. The RBD region is a critical target for nAbs for preventive as well as blocking therapy with antibodies. Further, the role of the S protein in infectivity also makes it an important tool for diagnostic antigen-based testing and vaccine development [15]. On the other hand, several monoclonal antibodies (mAbs) have shown promising results in neutralizing SARS-CoV-2. CR3022, a SARS-CoV-specific human mAb, binds potently with SARS-CoV-2, and has potential as therapeutic as well as prophylactic agent alone or in combination with other nAbs, for SARS-CoV-2 infection. In addition, sera from SARS patients during convalescence or animals specifically immunized with SARS-CoV S1 may cross-neutralize SARS-CoV-2 and reduce viral load or S protein-mediated SARS-CoV-2 entry.
Yuan, et al. have determined the structure of CR3022, the neutralizing antibody obtained from convalescent COVID-19 patients in complex with the receptor-binding domain of SARS-CoV-2 spike [16]. While Yu, et al. designed a series of prototype DNA vaccines against the SARS-CoV-2 S protein [17]. The analysis of the vaccine candidates in rhesus macaques have shown that the animals developed protective humoral as well as cellular immune responses when challenged with the virus. Neutralizing antibody titers were also observed at levels similar to those seen in humans who have recovered from SARS-CoV-2 infection [18].
In addition, the peptide fusion inhibitors such as nelfinavir mesylate, suppress both SARS-CoV-2 S and SARS-CoV S-mediated the virion-cell fusion. Nelfinavir may also inhibit TMPRSS2 involved in activation of the S protein. Camostat mesylate, a protease inhibitor targeting SARS-CoV-2 S cleavage sites, is a potent serine protease inhibitor of TMPRSS2 and appear to block the SARS-CoV-2 cellular entry [19].
The COVID-19 pandemic has dramatically accelerated global vaccine development efforts, most targeting the viral S protein. There are SARS-CoV-2 vaccine candidates based on types DNA- and RNA-based formulations, recombinant subunits containing viral epitopes, adenovirus-based vectors, and inactivated virus, under development. The inactivated virus vaccines have been traditionally used. Thus, inactivated virus vaccine development for SAR-CoV-2 may be a time-tested and effective way for the prevention of disease [20]. Gao, et al.
developed experimentally a purified inactivated SARS-CoV-2 virus vaccine candidate, PiCoVacc, which was able to induce SARS-CoV-2–specific neutralizing antibodies in mice, rats, and nonhuman primates. These antibodies neutralized various SARS-CoV-2 strains, thus displaying a possible broader neutralizing ability. The animals were immunized with one of two vaccine doses and then inoculated with SARS-CoV-2. Those having received the lowest dose showed signs of controlling the infection, whereas those receiving the higher dose appeared more protected with undetectable viral load in the pharynx or lungs at 7 days after infection.
The immune response to S protein: In the early phase of SARS-CoV-2 infection, T- and B-cell counts are substantially decreased [21]. In a period of about 2 weeks following onset of symptoms, IgM and IgG become detectable. The T cells play an important role in supporting the development of the B cell response, while the B cells produce the antibodies that recognize SARS-CoV-2 S protein. A particular subset of T cells, called T-follicular helper (TFH) cells, is predictor of an effective immune response. The circulating TFH cells are S-specific and functional, and the occurrence of CXCR3+ TFH cells is positively correlated with neutralising antibody titres in COVID-19-convalescent individuals [22].
The S glycoprotein plays essential roles in virus attachment, fusion, and entry into the host cell. Simultaneously, its surface location renders it a directly accessible target for host immune response [23]. The SARS-CoV-2 RBD has a high hACE2 binding affinity than SARS-CoV to support efficient cell entry. Further, SARS-CoV-2 RBD is less exposed than SARS-CoV RBD and helps the virus to evade immune response. Furthermore, the SARS-CoV-2 spike contains a proprotein convertase (PPC) motif at the S1/S2 boundary and its prior cleavage during viral packaging enhances the efficiency for entry into new target cells including those with low expression of TMPRSS2 and other proteases [24].
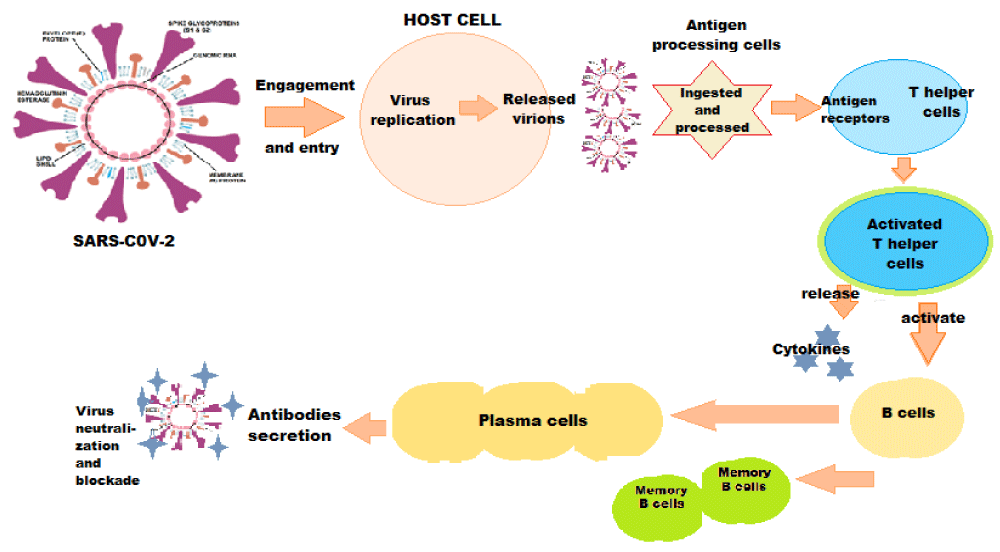
The immune system responds to the S protein, both S1 and S2 subunits being highly antigenic [25]. As a result, the S-specific antibodies, memory B and circulating TFH cells are consistently elicited following SARS-CoV-2 infection and COVID-19 vaccine shots in the clinical trials (Figure 2).
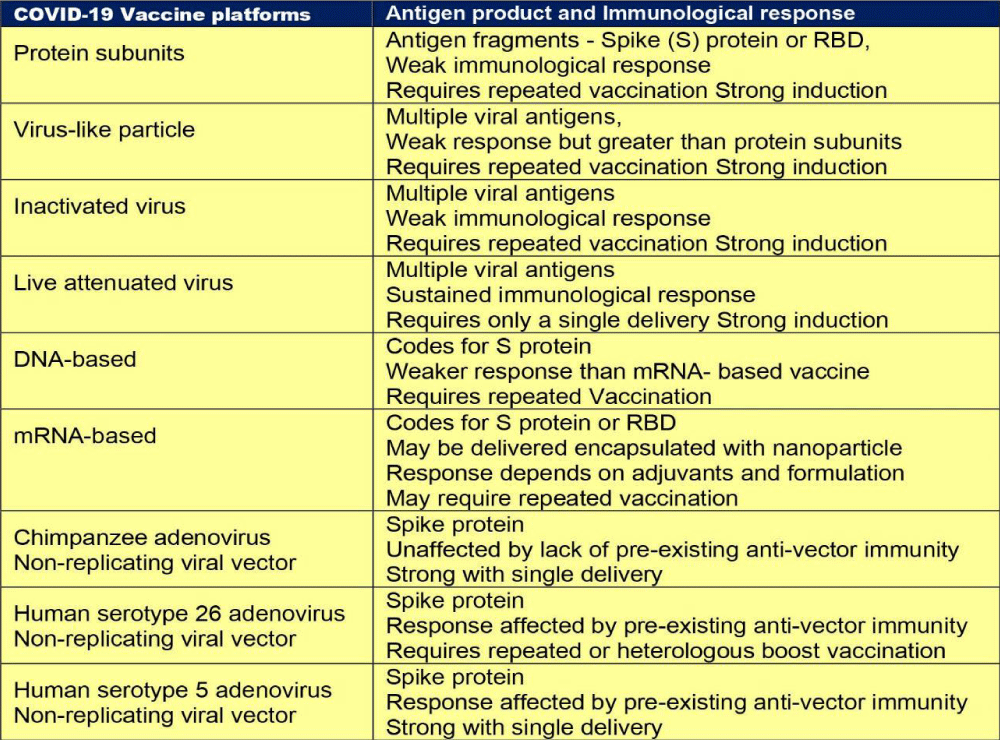
As reported during September 2021, there were nine different technology platforms (Table 1) under research and development to create an effective vaccine against COVID‑19 [22].
Figure 2: Immune Response against SARS-CoV-2.
There evolves humoral and circulating TFH immunity against S protein in the convalescent COVID-19 patients and those with robust B cell response show a strong plasma neutralising activity [26]. Still, in the COVID-19-convalescent individuals, the titres of spike-specific neutralising antibodies are variable [27]. In the clinical studies, the convalescent individuals who experienced severe COVID-19 showed higher neutralising antibody titres, a faster increase in lymphocyte counts and a higher frequency of CXCR3+ T follicular help (TFH) cells compared with COVID-19-convalescent individuals who experienced non-severe disease [28]. Remarkably, those recovering from severe COVID-19 elicit and maintain higher antibody and neutralization titres than the non-severe group. Thus, the neutralising antibody titre appears to correlate with the severity of the disease [29].
These findings provide insights into neutralising antibody responses in COVID-19-convalescent individuals and may facilitate the treatment and vaccine development for SARS-CoV-2 infection. The neutralising antibodies are crucial in protecting from re-infection. They bind to the S protein as well as prevent it from getting to attached to the host cells to facilitate virus entry [30]. The presence of anti-spike or anti-nucleocapsid IgG antibodies has been associated with a substantially reduced risk of SARS-CoV-2 reinfection in the ensuing 6 months [31]. Thus, generating a strong neutralising antibody response is the primary goal for SARS-CoV-2 vaccines.
Associated immunological phenomena: Antibodies predating COVID-19 infection: It has been documented that immunological memory after infection with seasonal human coronaviruses (hCoVs) may potentially contribute to cross-protection against severe acute SARS-CoV-2 infection [32]. It has been seen that a proportion of SARS-CoV-2–uninfected persons have circulating immunoglobulin G (IgG) antibodies that could cross-react with the S2 subunit of the SARS-CoV-2 S protein. By contrast, the COVID-19 patients generate IgA, IgG, and IgM antibodies that recognize both the S1 and S2 subunits.
Experimentally, these anti-S2 antibodies from SARS-CoV-2–uninfected patients have neutralizing activity against both SARS-CoV-2 and SARS-CoV-2 S pseudo-types. A much higher percentage of SARS-CoV-2–uninfected children and adolescents show these antibodies compared with adults, which may be because children and adolescents generally have higher hCoV infection rates and a more diverse antibody repertoire, which may explain the age distribution of COVID-19 susceptibility [33].
Similarly, the study by Demers-Mathieu et al. has evaluated the presence and the levels of antibodies reactive to SARS-CoV-2 S1 and S2 subunits in the human milk. These antibodies are reactive to S protein and nucleocapsid protein and could provide passive immunity to breastfed infants and protect from COVID-19 [34].
There exists striking structural similarity and sequence conservation between the SARS-CoV-2 S and SARS-CoV S glycoproteins that recognize hACE2 to enter the host cells. This resemblance is further strengthened by finding that SARS-CoV S elicits polyclonal antibody (pAb) response which can potently neutralize the SARS-CoV-2 S-mediated entry into the cells. Most of the SARS-CoV neutralising Abs target the SB domain and several of them recognize the RBM and prevent receptor engagement [35].
Antibody dependent enhancement: The SARS-CoV-2 inactivated whole-virion vaccine candidates carry the risk of viral infection of Fc receptor (FcR)-expressing cells, a phenomenon called antibody-dependent enhancement (ADE). Similarly, all other SARS-CoV-2 vaccine candidates entering clinical trials contain or express full-length or near full-length S protein and therefore also bear risk of ADE, albeit less so [36]. Considering this phenomenon, the continued search for a safe and effective SARS-CoV-2 vaccine may appear a farther dream but should go on.
The recombinant RBD proteins of SARS-CoV and MERS-CoV have been shown to potently induce protective nAbs. Further, it has been shown that the RBD part within RBD-Fc fusion protein is responsible for induction of nAbs against SARS-CoV-2. Furthermore, the anti-SARS2-RBD sera have been found to cross-react with SARS-RBD. Therefore, the observed cross-binding with SARS-RBD and cross-neutralization of SARS-CoV is likely to be contributed by antibodies targeting the conserved SARS-CoV-2 core subdomain, which contains cross-neutralization antibody epitopes. Zang, et al. have documented that the anti-RBD sera exhibited potent neutralization effects on SARS-CoV-2. Moreover, the anti-RBD sera also inhibited SARS2-S-mediated cell–cell fusion. On this basis, it seems that the anti-RBD antibodies do not promote ADE [37].
Development of COVID-19 vaccines: Various SARS-CoV-2 vaccine candidates based on various vaccine platforms, such as inactivated or live attenuated vaccines, DNA and mRNA vaccines, viral vector-based vaccines, and recombinant protein-based vaccines, have been developed [38]. Most of these vaccine strategies are based on the full-length S glycoprotein, which is the major SARS-CoV-2 surface antigen. Though the S protein is thought to be a promising vaccine immunogen for generating protective immunity, optimizing antigen design is critical to ensure an optimal immune response through exposing more neutralising epitopes and displaying fewer potentially weakly or non-neutralizing epitopes.
1. There is fear of erratic immune response to the S protein-based vaccines [39]. The RBM appears the most immunodominant neutralizing epitope of the whole S protein, capable of readily eliciting strong neutralizing antibody response but presents difficulties in designing and developing vaccine because of certain issues as folloThe native trimeric SARS-CoV-2 S protein conceals its immunodominant RBMs by adopting the closed conformation. The SARS-CoV-2 evades immune surveillance also through conformational masking [40].
2. The S1 subunit spontaneously dissociates from the S glycoprotein as a trimer to assume the RBD closed conformation, leaving only the post-fusion S2 trimer. The resulting S1 and S2 subunits might expose immunodominant, non-neutralising epitopes that are utilized by SARS-CoV-2 to serve as decoys to distract the host immune system, inducing a large proportion of ineffective antibody responses.
3. Further, the vaccine candidates based on the full-length S protein of the closely related SARS-CoV could elicit neutralising antibody responses against infection of SARS-CoV, which may also induce harmful immune responses, including liver damage of the vaccinated person, infection of human immune cells by SARS-CoV, and antibody-dependent enhancement of SARS-CoV infection.
Besides the RBD, which has been shown to be a major target for human neutralising antibody responses, the NTD has recently been identified to be a new vulnerable site of the SARS-CoV-2 S protein for antibody mediated neutralization and therefore could also serve as a recombinant protein-based vaccine [41]. The NTD-specific neutralising antibodies are likely to target the S protein in both closed and open conformations. In addition, the apparent accessibility of the FP and HR1 region in the SARS-CoV-2 S ectodomain trimer may be good immunogen candidates for epitope-focused vaccine design aimed at raising effective CoV neutralising antibodies.
Emerging SARS-CoV-2 strains
Mutations and variation in SARS-CoV-2: Understanding the nucleotide variations in the SARS-CoV-2 genome provides a useful insight for the evolution of the disease and the propagation of the pandemic [42]. The early variations in SARS-CoV-2 have made their way almost unnoticed as the virus spread around the world. Whereas most variations or mutations have no impact on the viral ability to transmit or cause disease, some mutations appear to have impact on transmissibility, infectivity, or lethality. Some of these mutations have possibly arisen because of the virus evolving from immune selection pressure in infected individuals and are more prevalent in patients with mild than those with severe disease. In general, the mutations can be interpreted as a strategy through natural selection to facilitate extensive spread of the viral infection.
Though, the SARS-CoV-2 virus has a low mutation rate by virtue of the nsp14 protein acting as a 3′-5′ exoribonuclease on both single-stranded and double-stranded RNA during the viral replication cycle [43]. Still its large genome appears to facilitate recombination, insertions, and deletions. Andrés et al found that the viral S protein accumulates deletions upstream and close to the S1/S2 cleavage site [44]. Further, SARS-CoV-2 can resort to RNA viral evolution through recombination (synthesis of chimeric RNA molecules from two different progeny genomes) and reassortment (the packaging within a single virion of genomic segments from different progeny viruses.
The single nucleotide variations (SNVs) as SARS-CoV-2 Spike amino acid replacements in the receptor binding domain (RBD) occur relatively frequently [45]. There is recurrent emergence and significant onward transmission of a six-nucleotide deletion in the Spike gene resulting in loss of two amino acids labelled as ΔH69/ΔV70. This deletion often co-occurs with the receptor binding motif amino acid replacements N501Y, N439K and Y453F. As such, these deletions have been found in a small percentage (2.2%) of the samples [46]. Currently the major SARS-CoV-2 Lineages are A, B, B.1, B.1.1, B.1.177. The lineage - B.1.1.7, of present concern, was first sequenced on 20 Sep 2020 and is spreading from the UK and has been discovered in Denmark, the Netherlands, Italy, Israel, Australia, Hong Kong, and Singapore. Other countries are being increasingly involved. Taking a note of the major mutations, their lineages and effects on disease transmissibility is important to understand the changing face of the pandemic (Table 1).
Table 1: Major mutations, lineages and effects on disease transmissibility and course.
Using the complete sequences of 1,932 SARS-CoV-2 genomes, six types of the strains have been identified. The 13 signature variations in the form of SNVs in protein coding regions and one SNV in the 5′ untranslated region (UTR) provide interpretation for the six types (types I to VI). The type VI, characterized by the four signature SNVs C241T (5′UTR), C3037T (nsp3 F924F), C14408T (nsp12 P4715L), and A23403G (Spike D614G), with strong allelic associations, first reported in China, has become the dominant type world over. Out of these, C241T is in the 5′ UTR appears to be of uncertain significance. The other three SNVs, 3037T-14408T-23403G characterizing the increasing frequency of the type VI, in majority of samples from various regions suggests a possible fitness gain for the virus. Further, the strains missing one or two of these signature SNVs fail to persist or wiped out by the evolutionary more fit variants [47].
Emergence of the D614G variant: The genome analysis of various isolates of the SARS-CoV-2 shows several regions having an increased proportion of some variants. One such variant is the D614G mutation in the C-terminal end of the S1 domain and in proximity to the S2 subunit. This variant has increased in prevalence with rapidity, becoming the predominant variant now world-over [48]. The D614G variant is associated with the faster viral transmission and harbouring and discharge of higher viral loads [49]. The variant has been found to be associated with increased Infectivity and reduced S1 Shedding [50]. Thus, the presence of glycine at 614 appears to improve the S-Protein Stability and Increase its incorporation into virions. Further, the structural analyses have revealed that the RBD of the D614G form of the spike protein is more likely to assume an “open” conformation than the RBD of the ancestral D614S form, implying an improved ability to bind to the hACE2 receptor. Furthermore, the higher viral load with D614G is consistent with epidemiological data suggesting enhanced infectivity associated with D614G [51].
The studies in hamsters infected with D614S or D614G variants, Plante, et al. have documented that the contemporary D614G variant replicated to higher titers in nasal-wash samples early after infection and outcompeted the ancestral D614S variant. These findings suggest increased viral fitness for D614G in a major upper airway compartment potentially associated with enhanced transmission [52]. The D614G variant was found to be equally sensitive to neutralizing antibodies and did not cause more severe disease than the ancestral strain in hamsters, a finding that supports current findings in humans [53].
Emergence of VUI-202012/01 variant: Another new variant, named VUI-202012/01 (Variant under Investigation, year 2020, month 12, variant 01), has been identified through viral genomic sequencing in the United Kingdom (UK). It has 17 mutations that may lead to a conformational change in the shape of the virus including the S protein. There is an N501Y mutation in the S protein, related to part alteration in the receptor-binding domain (RBD) and may result in the virus becoming more infectious. There are multiple spike protein mutations such as a double deletion (positions 69 and 70), deletion 144, N501Y, A570D, D614G, Y453F, P681H, T716I, S982A, D1118H) as well as mutations in other genomic regions. The preliminary analyses in the UK suggests that the UK variant B.1.1.7 has multiple mutations on it and is significantly more transmissible than previously circulating variants, with an estimated potential to increase the reproductive number (R) by 0.4 or greater with an estimated increased transmissibility of up to 70percent [54].
The variant was first detected during October 2020 in the UK from a sample taken in Sep 2020. The backwards tracing using genetic evidence suggests this new variant first emerged in September 2020 and then circulated at low levels in the population until mid-November, and thereafter it has spread alarmingly by mid-December [55]. It has been correlated with a significant increase in the rate of COVID-19 infection in the UK. The cases with the new variant have been from Denmark, Netherlands, and Belgium. The VOC 202012/01 variant has not been identified so far in the United States [56].
The B.1.1.7 variant has acquired 17 changes - 14 non-synonymous (amino acid [AA] altering) mutations, 6 synonymous (non-AA altering), and 3 deletions. The deletions include 69/70 deletion and P681H - near the S1/S2 furin cleavage site, in the S protein, and ORF8 stop codon (Q27stop) - in open reading frame 8. As reported by the UK, there is no clear epidemiological link to animals for VUI 202012/01 [57].
The emergence of VUI-202012/01 has certain likely implications:
• The probability of a wider spread of the new virus variant across the European nations and continents
• The potential impact on SARS-CoV-2 diagnostics - The UK reports that the deletion 69-70 in the spike protein of the variant causes a negative result from S-gene RT-PCR assays applied in some laboratories in the UK. This specific mutation has occurred many times in different countries and is geographically widespread.
• The potential impact on severity of disease in a population or groups - Potential impact on severity of disease in a population or group - The available information regarding severity of the new virus variant is limited. To date, there is no indication of increased infection severity observed related to the variant, but the assessment is challenged by the fact that the majority of cases were reported in people under 60 years old, who are less likely to develop severe symptoms.
• Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the UK. None of the previously described SARS-CoV-2 variants have been shown to cause increased infection severity; on the contrary, a clade 19B variant with lower severity was detected in Singapore in the spring and then disappeared.
• Frequency of reinfections - The mutations observed in the new variant are related to the receptor binding site and other surface structures, which may alter the antigenic properties of the virus. Based on the number and location of spike protein mutations, it seems likely that some reduction in neutralization by antibodies will be seen, but there is as yet no evidence that there is a resulting impact on increased risk for reinfection or lower vaccine effectiveness.
• The potential impact on vaccine effectiveness is a worrisome issue. It is being conjectured that the efficacy COVID-19 vaccines will remain like the ancestral SSARS-CoV-2 virus.
Recently a distinct phylogenetic cluster (named lineage B.1.1.7) was detected within the COG-UK surveillance dataset. This cluster has been growing rapidly over the past 4 weeks and since been observed in other UK locations, indicating further spread [58]. The B.1.1.7 lineage accounts for an increasing proportion of cases in parts of England. The number of B.1.1.7 cases, and the number of regions reporting B.1.1.7 infections, are growing. B.1.1.7 has an unusually large number of genetic changes, particularly in the spike protein. Three of these mutations - mutation N501Y in the receptor-binding domain (RBD) leads to increased binding affinity to human ACE2; the spike deletion 69-70del has been associated with evasion to immune response; and mutation P681H is immediately adjacent to the furin cleavage site.
Other isolated mutations
P681H mutation: The P681H is S protein mutation and involves the S1/S2 furin cleavage site. It has also emerged spontaneously multiple times and has been recently reported from Nigeria. There is no evidence to indicate that the P681H variant is contributing to increased transmission of the virus in Nigeria [59]. Earlier the analysis of samples collected in Aug 2020 and sequenced at the African Centre of Excellence for Genomics of Infectious Diseases (ACEGID), Nigeria, identified two SARS-CoV-2 sequences that share one non-synonymous SNP in S protein in common with this lineage. The non-synonymous SNV, S:P681H, has been observed in global data outside of the B.1.1.7 lineage.
At the moment, only about 1% of the SARS-CoV-2 genomes from Nigeria share any of the 17 protein-altering variants from the UK lineage of concern (B.1.1.7). Other reported mutations such as the N501Y, A570D, and the HV 69 - 70 deletion in S protein have not been detected in Nigeria currently.
ORF8 stop codon (Q27stop): This mutation is not in the spike protein but involves the open reading frame 8. Similar mutations have occurred in the past. In Singapore, one strain with this type of mutation emerged and disappeared. This is in line with the understanding that SARS-CoV-2 ORF8 is poorly conserved among coronaviruses. Among accessory genes, open reading frame 8 (ORF8) stands out by being highly variable and may be related with the virus ability to spread [60].
The pandemic and unmet challenges
COVID-19 as a disease and SARS-CoV-2 as its causative organism, continue to remain an enigma. While we continue to explore the agent factors, disease transmission dynamics, pathogenesis and clinical spectrum of the disease, and therapeutic modalities, the grievous nature of the disease has led to emergency and non-emergency authorizations for COVID-19 vaccines in various countries around the world.
Challenges posed by virus mutants: The genome data outlines that two SARS-CoV-2 virus samples collected from anywhere in the world differ by an average of just 10 RNA letters out of 29,903 [61]. Thus, there is a sluggish mutation rate. Despite this, through extensive research, researchers have catalogued more than 12,000 mutations in SARS-CoV-2 genomes. Many mutations appear not to affect the virus’s ability to spread or cause disease. In fact, the mutations altering the viral structural proteins are more likely to harm a virus and make it extinct rather than improve it. Further, various SARS-CoV-2 strains have no major impact but might in future on the course of the disease and pandemic, as they accumulate. Even an insignificant mutation can have a profound by downgrading the immune response.
The D614G mutation leads to favour open conformations and the viral entry into the host cells. Further, this mutation It is often accompanied by three mutations in other parts of the SARS-CoV-2 genome. The variation increases density of the S protein and viral infectivity [62]. There has been a rapid spread of D614G which was first spotted in samples collected early in the course of the pandemic from China and Germany. It has become the dominant strain across the European continent, Australia, Canada, and parts of the United States, and predicted to involve rest of the world regions. It appears that D614G represents a more transmissible form of SARS-CoV-2, which has emerged as a product of natural selection [63]. There is fear that a similar situation may occur with the VUI-202012/01 or B.1.1.7 variant.
Ill-defined and short-lived immune response: The studies in mice, monkeys and humans that received one of the experimental RNA vaccines, produced antibodies that proved more potent at blocking G viruses than D viruses [64]. With the G strain now ubiquitous, these findings are encouraging. Though, the experience with HIV that mutates to elude various vaccines developed so far, makes the researchers wary of the potential of SARS-CoV-2 to evade immunity and antigenic responses. The historic trial with the common cold Corona vaccine in the late 1980s, should be mentioned in this context, which included 15 healthy people given nasal solution containing inactivated common cold coronavirus. The monitoring for one year followed by analyses documented that most of them became infected before developing an effective immune defense. So far, various SARS-CoV-2 strains do not stop the immune system from developing neutralizing antibodies that recognize the virus. Still, there is a possibility that the virus may acquire mutations that either evade the immune response or alter the susceptibility to antibodies and immune cells.
In the experimental studies using animals and cell cultures, along with the latest molecular techniques, and in small human clinical studies, the immune response to SARS-CoV-2 has been recognized [65]. But it is not certain whether the immunity will be effective or lasting. Whether a vaccine will provide adequate protection, whether those who have recovered from COVID-19 can return to pre-pandemic behaviours and how readily the world can reduce the threat posed by the disease. The reinfections have been shown to occur [66]. Neither the frequency nor the elements of the immune response associated with reinfection have been studied and well documented.
The future course of COVID-19 pandemic: The, virus SARS-CoV-2 is here to stay for the foreseeable future [67]. The future of the disease, of course, appear to depend on various unknown factors, which include the effects of seasonality on the disease transmission and spread, the degree and duration of effective immune response to the disease, and the life-style choices made by individuals and measures enforced by the governments. Further, the pandemic’s course will also depend on the availability of a COVID-19 vaccine, it’s effectively and duration of its protective immune response. Concerning the latter, the significance of viral mutations should also be borne in mind and highlighted. There are serious challenges posed by SARS-CoV-2 virus and COVID-19 as the disease to the humanity. Let us hope that the today’s unmet challenges are solved in near future.
- Nikhra V. COVID-19 pandemic, recurrent outbreaks, and prospects for assimilation of hCoV-19 into the human genome. Int J Clin Virol. 2020; 4: 111-115. https://www.clinvirologyjournal.com/journals/ijcv/ijcv-aid1029.php
- Pillay TS. Gene of the month: the 2019-nCoV/SARS-CoV-2 novel coronavirus spike protein. J Clin Pathol. 2020, 73:7, 366-369. PubMed: https://pubmed.ncbi.nlm.nih.gov/32376714/
- Lu R, Zhao X, Li J, Niu P, Yang B, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020; 395: 565–574. PubMed: https://pubmed.ncbi.nlm.nih.gov/32007145/
- Ou X, Liu Y, Lei X, Li P, Mi D, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020; 11: 1620. PubMed: https://pubmed.ncbi.nlm.nih.gov/32221306/
- Daly JL, Simonetti B, Klein K, Chen K, Williamson MK, et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020; 370: 6518; 861-865. PubMed: https://pubmed.ncbi.nlm.nih.gov/33082294/
- Walls AC, Park YJ, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020; 181: 281–292. PubMed: https://pubmed.ncbi.nlm.nih.gov/32155444/
- Huang Y, Yang C, Xu XF, Wall A, McGuire AT, et al. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020; 41: 1141–1149. PubMed:
- Wang Y, Liu M, Gao J. Enhanced receptor binding of SARS-CoV-2 through networks of hydrogen-bonding and hydrophobic interactions. PNAS. 2020; 117: 13967-13974. PubMed: https://pubmed.ncbi.nlm.nih.gov/32503918/
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, et al. Cryo-Em structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020; 367: 1260-1263. PubMed: https://pubmed.ncbi.nlm.nih.gov/32075877/
- Hussain M, Jabeen N, Raza F, Shabbir S, Baig AA, et al. Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J Med Virol. 2020; 92: 1580-1586. PubMed: https://pubmed.ncbi.nlm.nih.gov/32249956/
- Yan R, Zhang Y, Li Y, Xia L, Guo Y, et al. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020; 367: 6485: 1444-1448. PubMed: https://pubmed.ncbi.nlm.nih.gov/32132184/
- Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, et al. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020; 176: 104742. PubMed: https://pubmed.ncbi.nlm.nih.gov/32057769/
- Nguyen HL, Lan PD, Thai NQ, Nissley DA, O'Brien EP, et al. Does SARS-CoV-2 Bind to Human ACE2 More Strongly Than Does SARS-CoV? J Phys Chem B. 2020; 124: 7336–7347. PubMed: https://pubmed.ncbi.nlm.nih.gov/32790406/
- Hurtley SM. Another host factor for SARS-CoV-2. Science. 2020; 370: 805-807.
- Huang Y, Yang C, Xu XF, Xu W, Liu SW, et al. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacologica Sinica. 2020; 41: 1141-1149. PubMed: https://pubmed.ncbi.nlm.nih.gov/32747721/
- Yuan M, Wu NC, Zhu X, Lee CD, So RTY, et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020; 368: 630-633. PubMed: https://pubmed.ncbi.nlm.nih.gov/32245784/
- Yu J, Tostanoski LH, Peter L, Mercado NB, McMahan K, et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020; 369: 806-811. PubMed: https://pubmed.ncbi.nlm.nih.gov/32434945/
- Kelly PN. Prototype DNA vaccines for SARS-CoV-2. Science. 2020; 369: 783-785.
- Schütz D, Ruiz-Blancob YB, Münch J, Kirchhoff F, Sanchez-Garcia E, et al. Peptide and peptide-based inhibitors of SARS-CoV-2 entry. Adv Drug Deliv Rev. 2020; 167: 47-65. PubMed: https://pubmed.ncbi.nlm.nih.gov/33189768/
- Gao Q, Bao L, Mao H, Wang L, Xu K, et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020; 369: 77-81. PubMed: https://pubmed.ncbi.nlm.nih.gov/32376603/
- Wang F, Nie J, Wang H, Zhao Q, Xiong Y, et al. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J Infect Dis. 2020; 221: 1762–1769. PubMed: https://pubmed.ncbi.nlm.nih.gov/32227123/
- Ni L, Ye F, Cheng ML, Feng Y, Deng YQ, et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020; 52: 971–977. PubMed: https://pubmed.ncbi.nlm.nih.gov/32413330/
- Duan L, Zheng Q, Zhang H, Niu Y, Lou Y, et al. The SARS-CoV-2 Spike Glycoprotein Biosynthesis, Structure, Function, and Antigenicity: Implications for the Design of Spike-Based Vaccine Immunogens. Front Immunol. 2020; 11:576622. PubMed: https://pubmed.ncbi.nlm.nih.gov/33117378/
- Shang J, Wan Y, Luo C, Ye G, Geng Q, et al. Cell entry mechanisms of SARS-CoV-2. PNAS. 2020; 117: 11727-11734. PubMed: https://pubmed.ncbi.nlm.nih.gov/32376634/
- Shah VK, Firmal P, Alam A, Ganguly D, Chattopadhyay S. Overview of Immune Response During SARS-CoV-2 Infection: Lessons from the Past. Front Immunol. 2020; PubMed: https://pubmed.ncbi.nlm.nih.gov/32849654/
- Juno JA, Tan HX, Lee WS, Reynaldi A, Kelly HG, et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat Med. 2020; 26: 1428–1434. PubMed: https://pubmed.ncbi.nlm.nih.gov/32661393/
- Zhao J, Yuan Q, Wang H, Liu W, Liao X, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020; 71: 2027-2034. PubMed: https://pubmed.ncbi.nlm.nih.gov/32221519/
- Zhang J, Wu Q, Qu X, Wang Q, Wu J, et al. Spike-specific circulating T follicular helper cell and cross-neutralizing antibody responses in COVID-19-convalescent individuals. Nat Microbiol. 2020; 6: 51-58. PubMed: https://pubmed.ncbi.nlm.nih.gov/33199863/
- Chen G, Wu D, Guo W, Huang D, Wang H, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020; 130: 2620–2629. PubMed: https://pubmed.ncbi.nlm.nih.gov/32217835/
- Chen X, Li R, Pan Z, Qian C, Yang Y, et al. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell Mol Immunol. 2020; 17: 647–649. PubMed: https://pubmed.ncbi.nlm.nih.gov/32313207/
- Lumley SF, O’Donnell D, Stoesser NE, et al. Antibody Status and Incidence of SARS-CoV-2 Infection in Health Care Workers. N Engl J Med. 2020; NEJMoa2034545. PubMed: https://pubmed.ncbi.nlm.nih.gov/33369366/
- Klompus S, Leviatan S, Vogl T, et al. Cross-reactive antibody responses against SARS-CoV-2 and seasonal common cold coronaviruses. COVID-19 SARS-CoV-2 preprints from medRxiv and bioRxiv. 2020.
- Ng KW, Faulkner N, Cornish GH, Rosa A, Harvey R, et al. Pre-existing and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020; 370: 1339-1343. PubMed: https://pubmed.ncbi.nlm.nih.gov/33159009/
- Demers-Mathieu V, Do DM, Mathijssen GB, Sela DA, Seppo A, et al. Difference in levels of SARS-CoV-2 S1 and S2 subunits- and nucleocapsid protein-reactive SIgM/IgM, IgG and SIgA/IgA antibodies in human milk. J Perinatol. 2020; 1-10. PubMed: https://pubmed.ncbi.nlm.nih.gov/32873904/
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020; 181: 281-292. PubMed: https://pubmed.ncbi.nlm.nih.gov/32155444/
- Wen J, Cheng Y, Ling R, Dai Y, Huang B, et al. Antibody-dependent enhancement of coronavirus. Int J Infecti Dis. 2020; 100: 483-489. PubMed: https://pubmed.ncbi.nlm.nih.gov/32920233/
- Zang J, Gu C, Zhou B, Zhang C, Yang Y, et al. Immunization with the receptor-binding domain of SARS-CoV-2 elicits antibodies cross-neutralizing SARS-CoV-2 and SARS-CoV without antibody-dependent enhancement. Cell Discov. 2020: 6: 61. PubMed: https://pubmed.ncbi.nlm.nih.gov/32901211/
- Nikhra V. Stages in COVID-19 vaccine development: The Nemesis, the Hubris, and the Elpis. Int J Clin Virol. 2020; 4: 126-135. https://www.clinvirologyjournal.com/journals/ijcv/ijcv-aid1029.php
- Flanagan KL, Best E, Crawford NW, Giles M, Koirala A, et al. Progress and Pitfalls in the Quest for Effective SARS-CoV-2 (COVID-19) Vaccines. Front Immunol. 2020; 11:579250. PubMed: https://pubmed.ncbi.nlm.nih.gov/33123165/
- Cai Y, Zhang J, Xiao T, Peng H, Sterling SM, et al. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020; 369: 1586-1592. PubMed: https://pubmed.ncbi.nlm.nih.gov/32694201/
- Duan L, Zheng Q, Zhang H, Niu Y, Lou Y, et al. The SARS-CoV-2 Spike Glycoprotein Biosynthesis, Structure, Function, and Antigenicity: Implications for the Design of Spike-Based Vaccine Immunogens. Front Immunol. 2020; 11:576622. PubMed: https://pubmed.ncbi.nlm.nih.gov/33117378/
- Naqvi AAT, Fatima K, Mohammad T, Fatima U, Singh IK, et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim Biophys Acta Mol Basis Dis. 2020; 1866: 165878. PubMed: https://pubmed.ncbi.nlm.nih.gov/32544429/
- Robson F, Khan KS, Le TK, Paris C, Demirbag S, et al. Coronavirus RNA Proofreading: Molecular Basis and Therapeutic Targeting. Mol Cell. 2020; 79: 710-727. PubMed: https://pubmed.ncbi.nlm.nih.gov/32853546/
- Andrés C, Garcia-Cehic D, Gregori J, Piñana M, Rodriguez-Frias F, et al. Naturally occurring SARS-CoV-2 gene deletions close to the spike S1/S2 cleavage site in the viral quasispecies of COVID19 patients. Emerg Microbes Infect. 2020; 9: 1900-1911. PubMed: https://pubmed.ncbi.nlm.nih.gov/32752979/
- Kemp SA, Datir RP, Collier DA, Ferreira IATM, Carabelli A, et al. Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion ΔH69/ΔV70. bioRxiv preprint. 2020.
- Wang F, Huang S, Gao R. Zhou Y, Lai C, et al. Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discov. 2020; 6: 83. PubMed: https://pubmed.ncbi.nlm.nih.gov/33298875/
- Yang HC, Chen CV, Wang JH, Liao HC, Yang CT, et al. Analysis of genomic distributions of SARS-CoV-2 reveals a dominant strain type with strong allelic associations. PNAS. 2020: 117: 30679-30686. PubMed: https://pubmed.ncbi.nlm.nih.gov/33184173/
- Baric RS. Emergence of a Highly Fit SARS-CoV-2 Variant. N Engl J Med. 2020; 383: 2684-2686. PubMed: https://pubmed.ncbi.nlm.nih.gov/33326716/
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 Virus. Cell. 2020; 182: 812-827. PubMed: https://pubmed.ncbi.nlm.nih.gov/32697968/
- Zhang L, Jackson CB, Mou H, Ojha A, Rangarajan ES, et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv preprint. 2020. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7310631/
- Zhang L, Jackson CB, Mou H, Ojha A, Rangarajan ES,et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. Preprint. bioRxiv. 2020. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7310631/
- Plante JA, Liu Y, Liu J, Liu J, Xia H, et al. Spike mutation D614G alters SARS-CoV-2 fitness and neutralization susceptibility. Nature. 2020; rs.3.rs-70482. PubMed: https://pubmed.ncbi.nlm.nih.gov/32935091/
- Hou YJ, Chiba S, Halfmann P, Ehre C, Kuroda M, et al. SARS-CoV-2 D614G Variant Exhibits Enhanced Replication ex vivo and Earlier Transmission in vivo. Preprint. bioRxiv. 2020; 2020. 09.28.317685. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7536872/
- Emerging SARS-CoV-2 Variants In UK Like The B.1.1.7 Strain And The 501.V2 Strain In South Africa And Elsewhere Will Change Course Of COVID-19 Badly. 2020. https://www.biogenetech.co.th/wp-content/uploads/2020/12/2-Emerging.pdf
- Chand M, Hopkins S, Dabrera G. Investigation of novel SARS-COV-2 variant: Variant of Concern 202012/01 (Report). Public Health England. 2020.
- Washington N, White S, Barrett KS. for the Helix Research Team. Is the new SARS-CoV-2 UK variant (B.1.1.7) already in the US? Maybe. CDC on B.1.1.7 mutant. 2020. https://www.cdc.gov/coronavirus/2019-ncov/more/scientific-brief-emerging-variant.html
- European CDC on B.1.1.7 mutant: https://www.ecdc.europa.eu/sites/default/files/documents/SARS-CoV-2-variant-multiple-spike-protein-mutations-United-Kingdom.pdf
- Report Rambaut A, Loman N, Pybus O, et al. on behalf of COVID-19 Genomics Consortium UK (CoG-UK)9. Report – Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563.
- Nigeria identifies new variant of COVID-19 in samples collected in August: Report. Devdiscourse News Desk. 2020. https://www.devdiscourse.com/news-source/Devdiscourse%20News%20Desk
- Pereira F. Evolutionary dynamics of the SARS-CoV-2 ORF8 accessory gene. Infect Genet Evol. 2020; 85: 104525. PubMed: https://pubmed.ncbi.nlm.nih.gov/32890763/
- Mercatelli D, Giorgi FM. Geographic and Genomic Distribution of SARS-CoV-2 Mutations. Front Microbiol. 2020. PubMed: https://pubmed.ncbi.nlm.nih.gov/32793182/
- Zhang L, Jackson CB, Mou H, Ojha A, Peng H, et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun. 2020; 11: 6013. PubMed: https://pubmed.ncbi.nlm.nih.gov/33243994/
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020; 182: 812-827. PubMed: https://pubmed.ncbi.nlm.nih.gov/32697968/
- Poland GA, Ovsyannikova IG, Kennedy RBl. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020; 396: 1595-1606. PubMed: https://pubmed.ncbi.nlm.nih.gov/33065034/
- Florindo HF, Kleiner R, Vaskovich-Koubi D, Acúrcio RC, Carreira B, et al. Immune-mediated approaches against COVID-19. Nat Nanotechnol. 2020; 15: 630–645. PubMed: https://pubmed.ncbi.nlm.nih.gov/32661375/
- Tillett RL, Sevinsky JR, Hartley PD, Kerwin H, Crawford N, et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. The Lancet Infeft Dis. 2020; 21: 52-58. PubMed: https://pubmed.ncbi.nlm.nih.gov/33058797/
- Max Planck Institute for the Science of Human History. COVID-19 is here to stay for the foreseeable future: Future of field-based sciences in the time of coronavirus. ScienceDaily. 2020. www.sciencedaily.com/releases/2020/09/200914112206.htm