More Information
Submitted: 29 September 2020 | Approved: 10 October 2020 | Published: 12 October 2020
How to cite this article: Nikhra V. COVID-19 pandemic, recurrent outbreaks and prospects for assimilation of hCoV-19 into the human genome. Int J Clin Virol. 2020; 4: 111-115.
DOI: 10.29328/journal.ijcv.1001025
ORCiD: orcid.org/0000-0003-0859-5232
Copyright License: © 2020 Nikhra V. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: ACE-2 receptors; COVID-19 pandemic; Endogenous retroviruses (ERVs) ; Genetic factors; hCoV-19; Proofreading proteins; RNA viruses; SARSCoV-2; Transposons; VIPs
COVID-19 pandemic, recurrent outbreaks and prospects for assimilation of hCoV-19 into the human genome
Vinod Nikhra*
Senior Chief Medical Officer and Consultant, Department of Medicine, Hindu Rao Hospital and NDMC Medical College, New Delhi, India
*Address for Correspondence: Vinod Nikhra, Senior Chief Medical Offi cer and Consultant, Department of Medicine, Hindu Rao Hospital and NDMC Medical College, New Delhi, India, Email: [email protected]; drvinodnikhra@rediff mail.com
The outbreaks and resurgence: The disease which reportedly began in the Chinese city Wuhan in November-December 2019, soon spread to various parts of the world, and was named and declared a pandemic disease by WHO. While the European countries were recovering from the epidemic, the disease took hold in the USA, the South American countries, Arabian countries, and South Asian countries, predominantly affecting Brazil, Peru, Iran, and India. Presently, many European countries are witnessing a resurgence and recurrent outbreaks of COVID-19.
Spread and evolving new insights: Whereas there is workplace-related infection rise as people are returning to their offices, in other places the outbreaks are related to the people crowding and meeting care-freely and trying to resort back to their earlier way of life. The reopening of the educational facilities across the continents may make matters worse.
Impact on health and healthcare: Most cases of COVID-19 infections go unnoticed and are followed by self-recovery. But what may appear good from the clinical perspective, appears to complicate epidemiological efforts to contain the outbreak. With the evolving information about the disease, there seem to be certain possible outcomes such as control and containment, or the persistence of the disease as global endemic accompanied with outbreaks and resurgent episodes.
Gnetic factors linked to disease severity: With the COVID-19 pandemic, not all infected patients develop a severe respiratory illness. Further, there is a large variation in disease severity, which may be due to the genetic factors underlying the variable response to the virus. It is becoming clear that apart from the advanced age and pre-existing conditions, certain genetic constituent factors render some patients more vulnerable to the more severe forms of the diseases.
Integration of virus into human genome: A significant part of the human genome is derived from viruses especially the RNA viruses. In fact, about 8 percent of the human genome is made up of endogenous retroviruses (ERVs), which are viral gene sequences that have become a permanent part of the human lineage after they infected our ancient ancestors. With this background, a novel concept emerging that if COVID-19 persists for several generations, its genetic material is projected to be integrated or assimilated into human genome. The involved mechanisms are conceptualized through the transposons or transposable elements of the SARS-CoV-2.
The COVID-19 resurgence and recurrent outbreaks
The disease which reportedly began in Chinese city Wuhan in November-December 2019 manifesting as respiratory illness with upper respiratory symptoms and pneumonia-like illness, soon spread to various parts of the world, and was named and declared a pandemic disease by the World Health Organization. The disease soon spread to certain European countries. While these countries were recovering from the epidemic, the disease took hold in UK and USA, and the South American countries, Arabian countries, and South Asian countries, predominantly affecting Brazil, Peru, Iran, and India. Presently, most of the European countries are witnessing recurrent outbreaks and a resurgence of COVID-19, whereas the epidemic is not yet over in other countries. This calls for a need for balanced views about the preventive measures, renewed vigilance and new control strategies including the enforced lockdown measures specifically targeting clusters of outbreaks, which play a major role in the disease spread.
The rising number of cases in European countries, though, currently are not comparable to the peaks earlier during April- May 2020, is a matter of great concern. The inference from data from diagnostic testing appears to suggest that these countries might have relaxed the lockdown measures too early and too much [1]. In general, the people’s willingness to stay alert and stick to new rules about social distancing and mask wearing appears to have vanished too, rather than becoming part of their behaviour pattern, leading to evolving new areas of fresh resurgence and outbreaks.
Evolving new insights into COVID-19 viral spread
1. Whereas there is workplace-related infection rise as people are returning to their offices, in other places the outbreaks are related to the younger people crowding, meeting and partying care-freely and other people trying to resort back to their earlier ways of life. Because most of the new outbreaks involve the cases in younger age groups, fewer of them die. But it is only a matter of time before the elderly people are affected leading to higher mortality. Further, reopening the educational facilities across the continents may make matters worse.
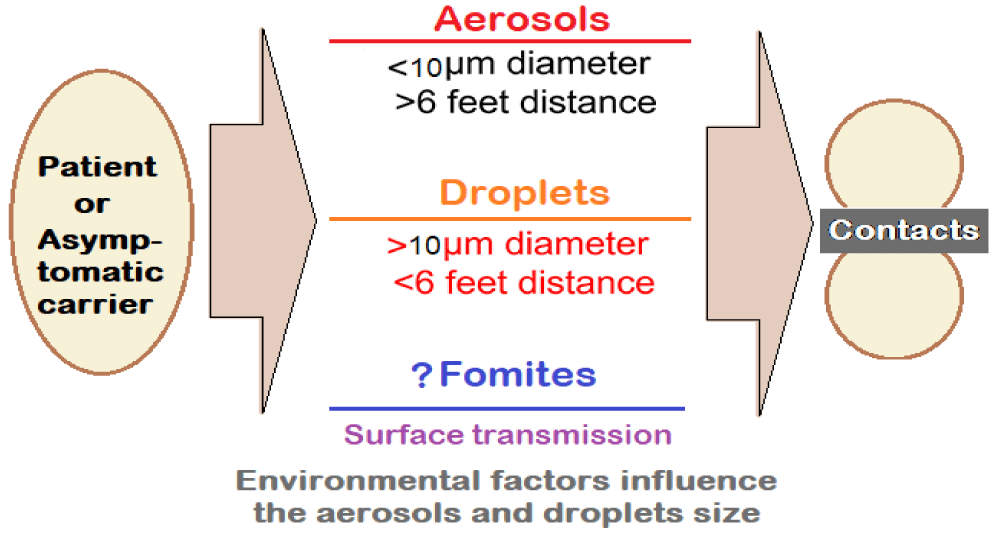
2. The airborne transmission: The transmissibility of SARS-CoV-2 is high. The infected individuals release droplets and aerosols containing SARS-CoV-2 by coughing or sneezing. The virus-laden aerosols and droplets can lead to short-range airborne transmission (~ 6 ft). The transmission may occur through fomites also but appears to play a minor role.
Much depends on the size of the aerosols (< 10-μm diameter) and droplets (> 10 μm diameter) which can promote infection through direct inhalation and deposition on surfaces and subsequent hand-to-mouth/nose/eye transfer (Figure 1).
Figure 1: Transmission kinetics for SARS-CoV-2: Droplets and Aerosol Concepts.
The suspended airborne droplets appear to persist in the air for several minutes. Whereas the smaller aerosols can persist for longer durations (several minutes to hours). The characteristics of aerosols are dynamic, due to evaporative loss of water depending on ventilation, and ambient humidity and temperature levels. With decreasing size, their ability to disperse in air is enhanced, leading to their transmission extending beyond 6 ft from the point of release.
3. The aerosol size fractions may influence the deposition profile of SARS-CoV-2 in the lung [2] Thus, whereas the larger aerosols (> 4 μm) are predominantly deposited in the upper and central airways (i.e., nasopharynx, tracheobronchial) and are subject to muco-ciliary clearance, the smaller aerosols (< 4 μm) get deposited deeper in the alveoli, having epithelial cells rich in ACE2, with enhanced transmission efficiency.
4. The emphasis on hand hygiene has been diluted, as it is becoming clear that contaminated surfaces may not play a large role. Similarly, the emphasis on banning outdoor activities is losing focus, as it is becoming established that the outdoor activities like jogging, outdoor hospitality, non-essential shopping, and public transportation are fine as long as people keep social distancing and wear face masks. Rather, the focus is on indoor activities which is the main culprit for the virus transmission.
5. The emphasis should now be on targeting outbreak clusters and super-spreading events. The studies indicate that 10% of patients cause 80% of all the infections, whereas most patients to the tune of 90% do not infect further [3]. The backward contact tracing is more useful than the forward tracing. Further, finding clusters help epidemiologists in understanding about the outbreaks. With more stress on the long-term care facilities and workplaces, the specifically targeted measures can help in preventing outbreaks, rather than general lockdown measures. These targeted versions include encouraging people to work from home and avoiding crowded places and banning meetings and gatherings.
6. Using the preventive measures and resources rationally, most of the countries are better prepared than before in the changed scenario. We are aware of the disease epidemiology, the virus transmission kinetics, and equipped with resources like PPE kits and masks. Further, the rational behaviour has emerged in place of irrational fears about the disease. The governmental organizations have developed machinery for contact tracing, surveillance and gathering data.
The impact of the disease on health and healthcare system
While most of COVID-19 infections go unnoticed and are followed by self-recovery. But what may appear good from the clinical perspective, vastly complicates epidemiological efforts to contain the disease outbreaks. With the evolving information about the COVID-19 so far, there appear to be following three possible outcomes.
Scenario 1: Control and Containment: The disease will persist with regional outbreaks and low endemicity. There will persist to occur periodic resurgence. The asymptomatic transmissions will continue to occur with occasional outbreaks and development of epicentres, requiring timely measures to curtail further spread.
Scenario 2: Persistence as global endemic: The overall mortality among known 2019-nCoV cases is about 2%. It appears that about 20% of infected people suffer severe disease [4]. Between these two groups are a multitude of people with milder forms of the disease, diagnosed or undiagnosed, who may or may not seek medical care. Many may have no symptoms at all. Thus, the diagnosed cases of the disease form the visible part of the largely hidden iceberg (Figure 2).
Figure 2: The COVID-19: Epidemiological Iceberg.
Scenario 3: If the disease goes on to persist and afflict the human populations over several generations, there are anticipated prospects of assimilation of hCoV-19 into Human Genome.
Genetic factors in clinical manifestations of COVID-19
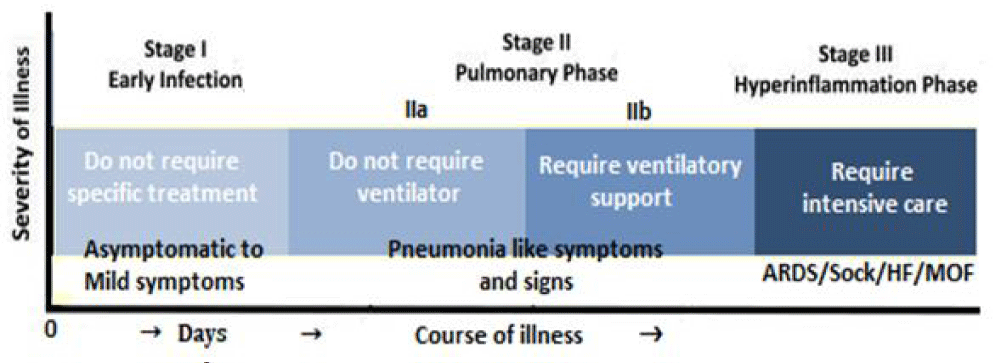
With the COVID-19 pandemic, not all infected patients develop a severe respiratory illness; the reason for which is not apparent. Further, it appears that there is a large variation in disease severity, one component of which may be due to the genetic variability in the response to the virus [5]. The individual response to SARS-CoV-2 exposure and the vulnerability of individuals to the infection, and the clinical spectrum of COVID-19 are greatly variable (Figure 3).
Figure 3: The clinical spectrum of COVID-19.
It is becoming clear that apart from the advanced age and pre-existing conditions, such as diabetes, cardiovascular, pulmonary, and renal diseases, certain genetic constituent factors render some patients more vulnerable to the more severe forms of the diseases, as is apparent from the rate of hospitalization of younger and apparently healthy individuals.
The host genetic factors have been linked to the variable clinical manifestations of the disease [6]. The clinical manifestations of COVID-19 patients have further been grouped based on (1) need for hospitalization, (2) need for oxygen supplementation, (3) progression to respiratory failure, or (4) mortality. From the clinical perspective, knowledge of host genetic factors could lead to improved care for patients with COVID-19. A model to understand human genomic variants linked to COVID-19 outcomes can be conceived as a continuum from ultrarare to common. Further, the genomic factors can be linked to variability in the protective immune response and have implications for vaccination strategies or could be used to optimally select patients for novel therapeutic treatments and trials.
The Genetic factors and Pathways related to COVID-19 involve -
The ACE2 gene encoding the ACE2 surface receptor associated with SARS-CoV-2 virus. The specific variants in the genes and pathways leads to difference in inter-individual COVID-19 susceptibility and response. The genetic polymorphisms in the ACE2 gene, which encodes the ACE2 receptors and allelic variants of the ACE2 may influence the virus binding subsequent to invasion of the cells.
Genes and pathways related to COVID-19 also include other viral receptor genes like related to TMPRSS2. The polymorphisms of cellular proteases, which facilitate the entry of SARS-CoV-2 into the cell, along with furin and TMPRSS2 - have been shown to exist. The TMPRSS2 variants and resulting expression may also influence COVID-19 severity, as well.
The course of the disease is also influenced by the inflammatory and immune response pathways such as IL-6 pathway, and genes involved in hypercoagulability and acute respiratory distress syndrome.
Other genes of interest include genes associated with ABO blood group [7]. A possible association between the genetic variability in histocompatibility complex (MHC) class I genes (human leukocyte antigen - HLA A, B, and C) and the susceptibility to SARS-CoV-2 and severity of COVID-19 has recently been suggested [8]. In Particular, the HLA-B*46:01 gene product is predicted to exhibit the lowest binding capacity to SARS-CoV-2 peptides and the individuals with this allele are more susceptible to COVID-19—due to reduced capacity for viral antigen presentation to immune cells. On the other hand, the HLA-B*15:03-encoded protein has the high capacity to present the conserved SARS-CoV-2 peptides that are shared among common human coronaviruses, leading the patients possessing this HLA genotype to more likely develop immunity.
Future prospects of COVID-19
Human genome and ERVs: A significant part of the human genome is derived from viruses [9]. In fact, the human genome is littered with various dormant viral genes [10]. About 8% of the human genome is made up of endogenous retroviruses (ERVs), which are viral gene sequences that have become permanently integrated parts of the human genome after infecting the human population groups during ancient times. The expression of these endogenous retroviruses has been implicated in diseases like autoimmune disorders and breast cancer. But they are also useful for human survival. For example, they play an important role as an interface between a pregnant mother and the fetus by regulating placental development and function.
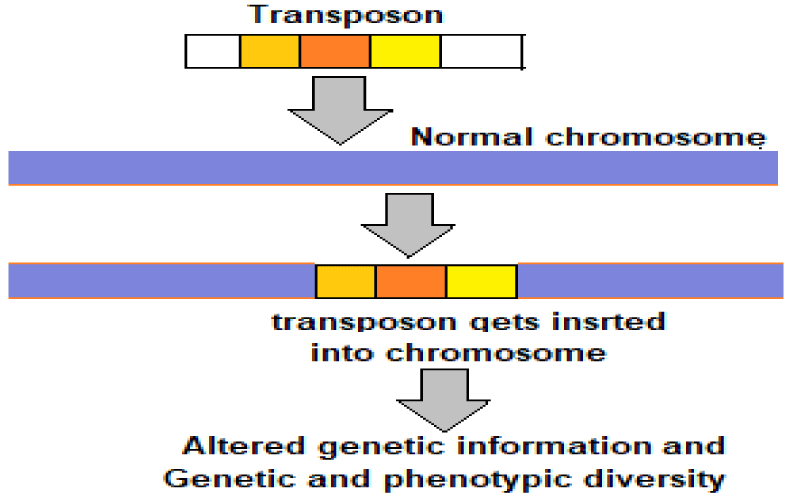
Transposable elements or transposons: Viruses are ancient and vital simplest genetic constructions. They tend to be made of a protective shell, a protein called a polymerase, responsible for replicating the viral genome, and a sequence of nucleotides — either RNA or DNA — that encodes for the viral proteins. A virus may exist in purely genetic form, lacking a defined body. A stimulus disturbing its dormancy, lets it rebuild the physical body from a purely genetic form. The physical body bestows means and necessary tools to replicate. The disembodied viruses are called transposable elements, or transposons, which are mobile genetic elements also called jumping genes, and can move in and out of genomes. Transposons are present in all life forms. They are often the main components of the moderately repetitive DNA. In human beings more than 50% genome is composed of mobile elements (MEs) or Transposons [11].
Due to their past incremental accumulation and ongoing DNA transposition, MEs serve as a significant source for both inter- and intra-species genetic and phenotypic diversity in the primate and human evolution. The transposons can copy and paste themselves throughout genomes (Figure 4). Some endogenous retroviruses (ERVs) are themselves transposons. As documented, nearly 8% of the human genome is made up of ERVs and nearly 50% of the human genome is made of transposons. The viruses through transposons are thought to play a major role in genetic change and influence the processes of evolution and speciation.
Figure 4: Transposons – the drivers of evolution.
The proteins interacting with viruses: Over the course of the last several million years of evolution, the ancient human-beings likely to have been plagued by a multitude of viral epidemics. In this context, the host genomes offer an indirect way to detect the ancient epidemics [12]. These pathogens have shaped the host genomes by driving large numbers of adaptations involving various genes. A past epidemic can be detected through the enrichment in signals of adaptation at the host proteins that interact with viruses, called virus interacting proteins or VIPs [13].
By using the enrichment in signals of adaptation at about 4,500 host loci that interact with specific types of viruses, there has been documented that RNA viruses have driven large number of adaptive events across diverse human populations. Various types of viruses appear to have exerted different selective pressures during human evolution through the host VIPs. Further, the interactions with viruses account for approximately 30% of protein adaptations in the human genome, which have driven human evolution through various viral epidemics in past. Further, the analysis of VIPs suggests that the certain specific viruses may have indeed driven more epidemics than others in recent evolution.
The novel zoonotic RNA viruses: The RNA viruses are potentially the most important group involved in zoonotic disease transmission. They have higher probabilities to infect new host species because of their exceptionally shorter generation times and faster evolutionary rates. As documented and highlighted, the RNA viruses are the most common class of pathogens responsible for new human diseases, with a rate of 2 to 3 novel viruses being discovered each year [14]. RNA viruses show remarkable capabilities to adapt to new environments and confront the different selective pressures they encounter including the host’s immune system and defense mechanisms.
The recent emergence of interspecies-transmitted RNA viruses, such as Chikungunya (CHIKV) and Zika (ZIKV) viruses, represent new global pandemics. Other zoonotic, highly communicable RNA viruses during recent years include Lassa fever, Ebolavirus, severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), and Influenza A virus (IAV) [15].
The proofreading proteins in CoVs: The peculiar rate of adaptive evolution of the RNA viruses arises from their exceptionally high mutation and substitution rates, with the RNA viruses showing greater substitution rates than DNA viruses. Further, most RNA viruses lack the proofreading ability of RNA Polymerase and Reverse transcriptase. A fallout of this is that in long run they lose infectivity as the number of non-viable viruses outnumber those viable ones. Whereas in the case of replicative DNA polymerases of DNA viruses and cellular organisms, an exonuclease corrects the possible nucleotide misincorporations during the genome replication.
The coronaviruses (CoVs), like the DNA viruses, have the potential proofreading functions as the nsp14 protein acts as a 3′-5′ exoribonuclease on both single-stranded and double-stranded RNA during the viral replication cycle [16]. Through this proof-reading function, the CoVs appear to have overcome the limitation, which in most RNA viruses, during process of replication, lead to the non-viable virions rapidly outnumbering the viable ones, leading to a loss of fitness and/or viral extinction. In addition, the CoVs, like other RNA viruses can resort to RNA viral evolution through recombination (synthesis of chimeric RNA molecules from two different progeny genomes) and reassortment (the packaging within a single virion of genomic segments from different progeny viruses) [17].
Projected sars-CoV-2 integration into human genome
Various salient features in structure and physiology of the SARS-CoV-2 make it a highly pathogenic RNA virus, which is likely to persist as infecting agent and can potentially afflict the human populations for innumerable years if not for many generations. With this background, an emerging novel concept holds that if COVID-19 persists for several generations, its genetic material is projected to be integrated or assimilated into human genome. The involved mechanisms have been conceptualised through the transposons or transposable elements of the SARS-CoV-2.
- Kupferschmidt K. Can Europe tame the pandemic's next wave? Science. 2020; 369: 1151-1152. PubMed: https://pubmed.ncbi.nlm.nih.gov/32883840/
- Tang S, Mao Y, Jones RM, Tan Q, Ji JS, et al. Aerosol transmission of SARS-CoV-2? Evidence, prevention and control. Environ Int. 2020; 106039. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7413047/
- Kupferschmidt K. Case clustering emerges as key pandemic puzzle. Science. 2020; 368: 808-809. PubMed: https://pubmed.ncbi.nlm.nih.gov/32439769/
- Kupferschmidt K, Cohen J. Will novel virus go pandemic or be contained? Science. 2020; 367: 610-611. PubMed: https://pubmed.ncbi.nlm.nih.gov/32029604/
- Pollitt KJG, Peccia J, Ko AI, Kaminski N, Dela Cruz CS, et al. COVID-19 vulnerability: the potential impact of genetic susceptibility and airborne transmission. Human Genomics. 2020; 14; 17. PubMed: https://pubmed.ncbi.nlm.nih.gov/32398162/
- Murray MF, Kenny EE, Ritchie MD, Rader DJ, Bale AE, et al. COVID-19 outcomes and the human genome. Genet Med. 2020; 22: 1175–1177. PubMed: https://pubmed.ncbi.nlm.nih.gov/32393819/
- Wu BB, Gu DZ, Yu JN, Yang J, Shen WQ. Association between ABO blood groups and COVID-19 infection, severity and demise: A systematic review and meta-analysis. Infect Genet Evol. 2020; 84: 104485. PubMed: https://pubmed.ncbi.nlm.nih.gov/32739464/
- Nguyen A, David JK, Maden SK, et al. Human Leukocyte Antigen Susceptibility Map for Severe Acute Respiratory Syndrome Coronavirus 2. J Virol. 2020; 94: e00510-20. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7307149/
- Moelling K, Broecker F. Viruses and Evolution - Viruses First? A Personal Perspective. Front Microbiol. 2019; 10: 523. PubMed: https://pubmed.ncbi.nlm.nih.gov/30941110/
- Callif BL. Organumics: An Epigenetic Re-framing of Life, Consciousness and Evolution. Kindle Edition, 2019 ASIN: B07ZMKMVSW, Publisher: S. Woodhouse Books, Everything Goes Media LLC.
- Tang W, Mun S, Joshi A, Han K, Liang P, et al. Mobile elements contribute to the uniqueness of human genome with 15,000 human-specific insertions and 14 Mbp sequence increase. DNA Res. 2018; 25: 5: 521-533. PubMed: https://pubmed.ncbi.nlm.nih.gov/30052927/
- Enard D, Petrov DA. Ancient RNA virus epidemics through the lens of recent adaptation in human genomes. 2020.
- Enard D, Cai L, Gwennap C, Petrov DA. Viruses are a dominant driver of protein adaptation in mammals. ELife. 2016; 5: e12469. PubMed: https://pubmed.ncbi.nlm.nih.gov/27187613/
- Rosenberg NA, Kang TL. Genetic Diversity and Societally Important Disparities. Genetics. 2015; 201: 1–12. PubMed: https://pubmed.ncbi.nlm.nih.gov/26354973/
- Carrasco-Hernandez R, Jácome R, Vidal YL, de León SP. Are RNA Viruses Candidate Agents for the Next Global Pandemic? A Review. ILAR J. 2017; 58: 343–358. PubMed: https://pubmed.ncbi.nlm.nih.gov/28985316/
- Robson F, Khan KS, Le TK, et al. Coronavirus RNA Proofreading: Molecular Basis and Therapeutic Targeting. Molecular Cell. 2020; 79: 710-727.
- Pérez-Losada M, Arenas M, Galáne JC, Bracho MA, Hillung J, et al. High-throughput sequencing (HTS) for the analysis of viral populations. Infect Genet Evol. 2020; 80: 104208. PubMed: https://pubmed.ncbi.nlm.nih.gov/32001386/