More Information
Submitted: May 27, 2025 | Approved: June 06, 2025 | Published: June 09, 2025
How to cite this article: Chowdhury SK, Mridha NK, Khan AA, Baildya N, Mandal M, Ghosh NN. COVID-19 Vaccines Development: Challenges and Future Perspective. Int J Clin Virol. 2025; 9(1): 010-020. Available from:
https://dx.doi.org/10.29328/journal.ijcv.1001064.
DOI: 10.29328/journal.ijcv.1001064
Copyright License: © 2025 Chowdhury SK, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Antigens; Covaxin; COVID-19 vaccine; Variants of mutant COVID-19; Immune response; mRNA vaccines; SARS-CoV-2
COVID-19 Vaccines Development: Challenges and Future Perspective
Swapan Kumar Chowdhury1, Niranjan Kumar Mridha2, Abdul Ashik Khan3, Nabajyoti Baildya4, Manab Mandal5* and Narendra Nath Ghosh6*
1Department of Botany, Balurghat College, Balurghat, West Bengal 733101, India
2Department of Chemistry, Gour Mahavidyalaya, Malda-732142, India
3Department of Chemistry, Darjeeling Government College, West Bengal 734104, India
4Department of Chemistry, University of Kalyani, Kalyani-741235, India
5Department of Botany, Dukhulal Nibaran Chandra College, Suti-742201, India
6Department of Chemistry, University of Gour Banga, Malda-732103, India
*Address for Correspondence: Dr. Manab Mandal, Assistant Professor, Department of Botany, Dukhulal Nibaran Chandra College, Suti-742201, India, Email: [email protected]
Narendra Nath Ghosh, Department of Chemistry, University of Gour Banga, Malda -732103, India, Email: [email protected]
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) outbursts began at the end of 2019, which imposed a serious crisis on public health and the economy all over the world. To date, there is no antiviral drug available for SARS-CoV-2, and hence vaccination is the most preferred method to prevent people from getting attacked by this virus, especially for those who are at high risk. To counter coronavirus-2, there are various types of vaccines, which are being used, such as live attenuated vaccines, killed or inactivated vaccines, recombinant vaccines, mRNA vaccines, recombinant vector vaccines, and DNA vaccines. Novavax data shows that the vaccine is effective against severe diseases caused by B.1.351. The Pfizer-BioNTech and AstraZeneca vaccines show evidence of some protection against P.1. Due to the immune response, the Human body can recognize and protect itself against harmful foreign substances such as bacteria, viruses, and microorganisms. The immune system protects our body from these harmful substances by identifying them as antigens. Virus-infected cells release many chemicals such as chemokines and cytokines for the initiation of immune response. To control the pandemic situation, herd immunity is required by the immunization of a critical mass of the world population at once. In this review article, we have made an analysis of the immune response of the human body to SARS-CoV-2 infection, different types, and modes of action of SARS-CoV-2 vaccines along with the current status of vaccines.
The human body can recognize and protect itself against foreign substances such as bacteria, viruses, and microorganisms that are harmful to us; this is called the immune response. The immune system protects our body from these harmful substances by identifying them as antigens [1,2]. Antigens (more specifically hetero-antigens) are mainly those proteins or fragments of protein that are present in the cell surface of fungi, viruses, and bacteria or produced by them. Nonliving substances may also act as antigens such as Chemicals, toxins, drugs, etc. Thus the immune system is how our body can recognize and destroy or tries to destroy, all of these types of pathogens (antigens) [3,4]. Mainly two types of immune responses are involved during viral infections in a host cell. The first one is the non-specific innate immune system which is also the 1st line of defense that determines the ‘molecular patterns’ of the entered pathogen [5-7]. Second, is the highly specific long-lasting memory effect of vig adaptive immune system [8]. Virus-infected cell releases many chemicals such as chemokines and cytokines for the initiation of immune response [9,10]. After that, neutrophils & macrophages are activated and thereby attracted to the virus-infected site and kill the virus-infected cell by releasing cytotoxic substances, such as matrix metalloproteinases [11,12].
In this mini-review we have made a comprehensive analysis of types of immune response systems, the kinds of vaccines developed so far and the proper mechanism of working of them against SARS-CoV-2.
Innate immune responses to SARS-CoV-2 infection
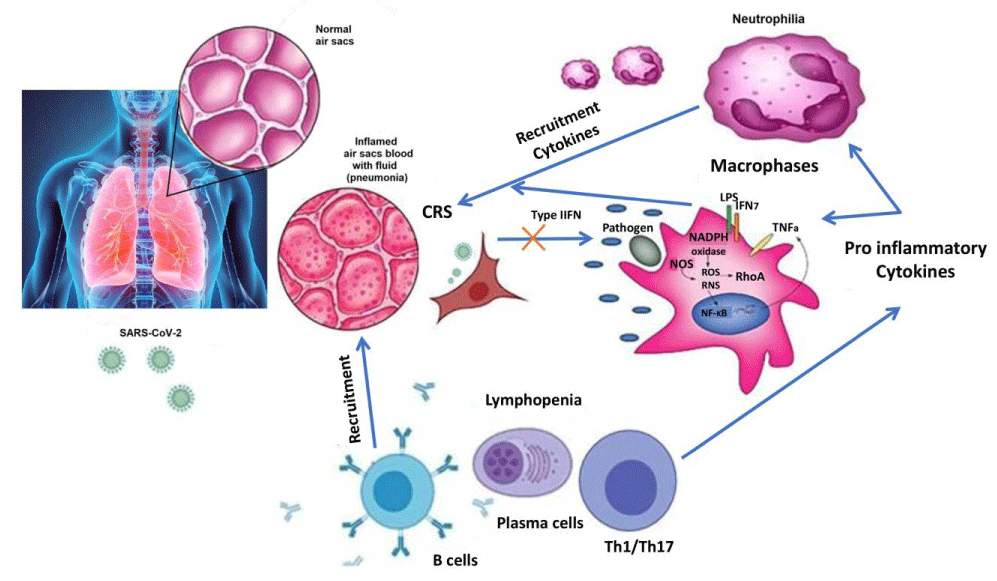
Pathogen-associated molecular patterns (PAMPs) molecules, lipopolysaccharide (LPS) are a diverse sets of microbial molecules that need to be recognized to elevate the antiviral response or innate immune system to destroy intruding pathogens [13-16]. These RNA base viruses including dsRNA are detected by the endosomal RNA receptor (TLR3 and TLR7) and the cytosolic RNA sensor (RIG-I/ MDA5) [17-19]. This process involves the activation of downstream signaling cascade (NF-κB and IRF3) along with nuclear translocation, which produces type I IFN expression, and pro-inflammatory cytokines to fight against pathogens at an early stage [20-22]. The innate immune response depends on the expression of IFN (IFN-alpha/beta) which are cytokines secreted from the virus-infected cell to the nearest healthy cell for its protection. Schneider, et al. reported that humans generate 13 subtypes of IFN-alphas, one IFN-beta, and one each of IFN-omega and IFN-kappa [23]. The signaling pathway is JAK-STAT which is completed by the binding of IFN-cytokines to the IFN-receptors (IFNAR-1 and IFNAR-2) [24-27]. Thereafter, Janus Kinases (JAK1 & JAK2) are activated followed by phosphorylating the signal transducers and transcription factors STAT1 and STAT2. Then IRF9 forms complexes with STAT1 & STAT2 and enters into the nucleus to initiate the transcription and finally up-regulated the gene expression. The total process of gene expression by IFN is called ISGs (IFN-stimulated genes) [28,29] and is shown in Figure 1. SARS-CoV infection suppresses the IFN signaling pathway and in turn, inhibits all the down-regulated signaling by the IFNAR [30-32]. This process also inhibits IRF3 nuclear translocation by the degradation of RNA sensors (RIG-i & MDA5) [33,34]. Therefore the JAK-STAT pathway is interrupted and phosphorylation or dimerization does not occur by STAT which also inhibits gene expression [35]. Both structural viral proteins (such as M, N) and non-structural viral proteins (ORF proteins) are involved in the modulation of this host type-I IFN response. From the genomic sequence comparison analysis, it is found that SARS-CoV-2 has approximately 79% similarity with previously occurred SARS-CoV and has been containing additional gene regions (10b, 13, 14). Apart from that 68% similarity has been observed between SARS-CoV and SARS-COV-2 from amino acid sequences. So the interference pattern of SARS-CoV-2 with the host innate immune response may be ascertained after careful genome sequence analysis. Although the mechanism of SARS-CoV-2 infection against innate immune response is still not clear, it may be hypothesized to some extent that SARS-CoV-2 has developed the same sort of approach to reduce and/or modulate the type-I IFN and the host innate immune response. Therefore, innate immune response plays a major role in the early detection of SARS-CoV infection by the secretion of chemokines and cytokines, especially type-I IFN from neutrophils and macrophages. The data analysis on the COVID-19-infected patients indicates that there is a similar change in total neutrophils and lymphocytes which is involved in delayed secretion of type I IFN. This ultimately leads to failure to control viral replication in an early phage. Young patients are comparatively less affected by COVID-19 due to their potential innate immunity system whereas old patients suffer severe conditions [36]. The mouse model study also supports the hypothesis that proper timing of innate immune response was responsible for protection against the RNA virus, COVID-19 [37].
The immune system plays a vital role in removing toxins and allergens that penetrate mucosal surfaces, relying on both innate and adaptive mechanisms that differentiate between self and nonself to effectively combat pathogens and other external threats, while also highlighting how disruptions in immune function can worsen tissue damage which shown in Figure 1.
Figure 1: Schematic pathway of innate immune response.
Adaptive immune responses
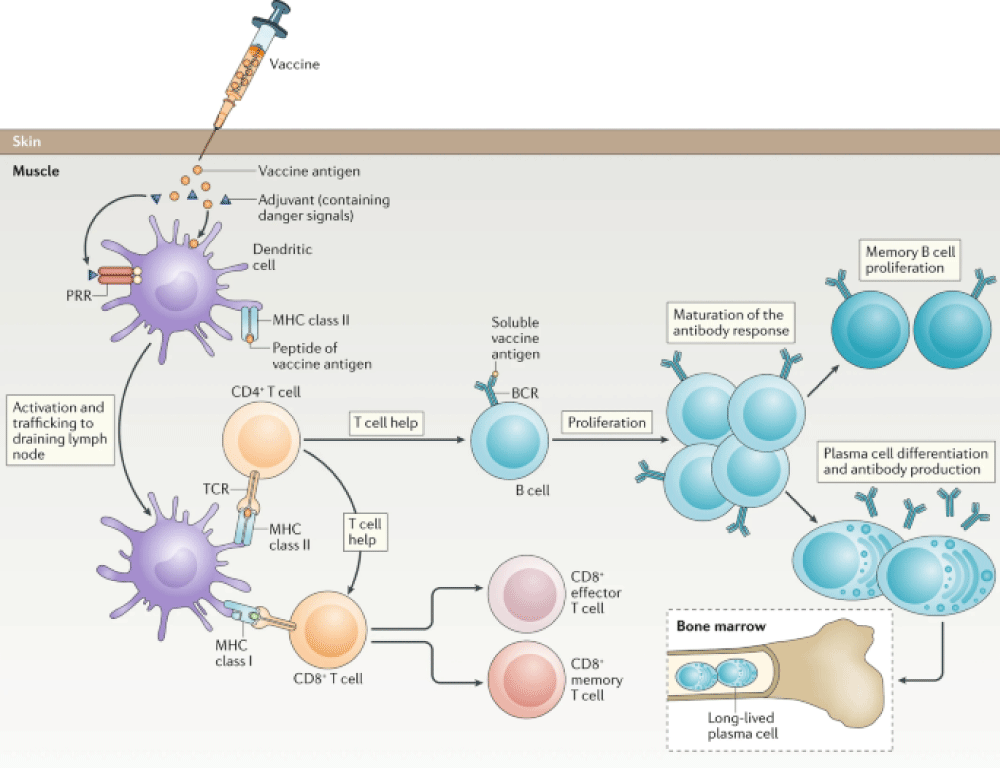
Adaptive immunity is highly specific to pathogens and only activated when the innate immunity system gives information about the foreign molecules via releasing several chemicals like interleukins from the affected macrophages and dendritic cells which form the MHC-II complex during the signaling pathway. The adaptive immune response is of two types: cell-mediated immune response (T cells) and humoral immune response (B cells and antibodies). T cells are of three types: cytotoxic, helper, and suppressor cells. Cytotoxic T cells can kill pathogens directly by secretion of IL-4 in the cell-mediated immune response and helper T cells activate B cells to produce antibodies in plasma cells and enhance the phagocytosis of pathogens [38]. Adaptive immunity provides information about the similar pathogen entered earlier by the memory cell to inhibit the re-infection quickly. There are two types of helper T cells involved (Th1 & Th2). Th1 secretes IL-10 & IL-2 to activate Th2 and Tc (Killer Cell) respectively. The humoral immune response involves B cell activation by TH2 through IL-4 signaling and produces antibodies to prevent re-infection in the future [39]. In SARS-CoV, both T and B cell epitopes were extensively mapped for the structural proteins, S, N, M, and E protein [40-42]. The serum analysis of convalescent SARS-infected patients showed that plasma contained antibodies which were mainly IgM and IgG. The SARS-specific IgG antibody persists for a long time up to 2 years after infection and acts as a humoral immune response, whereas the SARS-specific IgM antibody persists for a very short time (within 13 weeks). It was also considered that IgG antibody is highly specific to produce antibodies against the structural proteins (spike & nucleocapsid) of SARS-CoV [43-47]. Detailed report of serology of SARS CoV 2 is insufficient. A primary study for a patient shows that specific IgM after the onset infection of 9 days was very high and later on after 2 weeks changed to IgG [48,49]. The same study was carried out with 5 patients having COVID-19 positive which resulted in some cross-reaction with SARS-CoV only. In addition, a plaque assay indicated that patients likely produced antibodies to neutralize the SARS-CoV-2 virus, with recommendations about ways to boost the immune response. To identify the immune response that correlates with protection, Li, et al. studied T cell response against SARS-CoV infection. From the analysis of 128 convalescent samples of SARS-CoV patients, they observed that 90% controlled strongly neutralizing Abs and 50% were affirmative for T cell response [50]. CD4+ T cell responses were less significant as compared to CD8+ in terms of frequency and magnitude of the responses. CD4+ T cell responses were primarily huddled in spike protein but CD8+ T cell responses were spread extensively across the entire set of SARS proteins. Polychromatic cytometry analysis revealed that the largest component of virus-specific CD4+ T cells secreted only one cytokine, from CD4+ T cells in individuals with mild-to-moderate and severe SARS, prone to be central memory phenotype (CD27+/CD45RO+) formed IFN-gamma, TNF-α and IL-2 and CD8+ T cells also released IFN-gamma, TNF-α and degranulated state (CD107a) with significantly higher frequency as compared with mild-moderate group. Enriched TH2 cytokines (IL-4, IL-5, IL-10) were noticed, in case of severe SARS infection. It is clear from the present data that TH response might be a very effective tool to combat strongly against previous outbreaks of COVs (SARS and MERS) [51]. Thus the similar role of TH type response may help to prevent SARS-CoV-2. T cell’s response to infected cells is highly significant for the adaptive immune system. Based on the magnitude and frequency of response, CD8+ T cells response is more effective and significant as compared to CD4+ T cells as shown in Figure 2 [52]. Therefore, with the help of T cell’s immune response as a benchmark, the design, development, and evaluation of vaccines or the defense and eradication of CoVs are ascertained in the interest of future research. Structural proteins (S, E, M, and N) are the major constituents for the modeling of epitopes. Memory CD4+ T cells are guarded in an animal model for protection against CoVs and also neutrophils have a destructive role in any kind of infections [53]. A vaccine-mediated adaptive immune response involves several stages like antigen presentation, T cell and B cell activation, antibody production, and the formation of memory cells by introducing antigens, paving the way for lasting immunity against specific pathogens which is shown in Figure 2.
Figure 2: Schematic diagram of vaccine-mediated adaptive immune response.
What is vaccine?
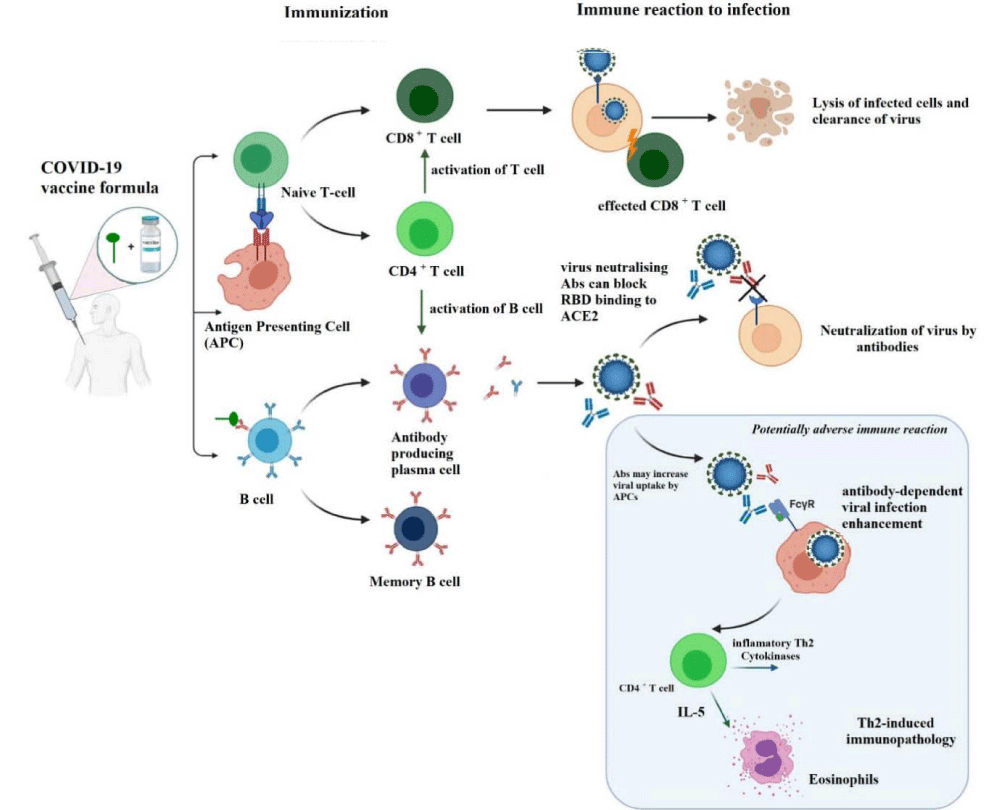
Vaccine is definitely a biological development that gives adaptive immunity against a specific contagious disease [54]. It may be produced by an agent that simulates with microorganisms accountable for disease or is often made from weakened/killed forms or it can originate from chemicals, toxins, and surface proteins [55]. To enhance the body’s immunity vaccines are introduced directly into the body via injection, mouth, or nasal route. The human body itself can generate antibodies against specific substances due to the defense mechanism that is obtained from the process of immunity development. Another important fact about vaccines is that it has a protective role against the effects of a future infection by a natural or “wild” pathogen or therapeutic to fight against a disease that has already happened [56].
Types of vaccines
There are various types of vaccines such as live attenuated vaccines, killed or inactivated vaccines, recombinant vaccines, mRNA vaccines, and recombinant vector vaccines as well as DNA vaccines. To date, there is no antiviral drug is available for SARS-Cov-2 and hence vaccination is the most preferred method to prevent people from this viral attack, especially for those who are at high risk due to compromised immunity or other health issues. So vaccination is the only option in our hands to prevent people from this coronavirus infection.
Live attenuated vaccines
The development of Live or attenuated vaccines can be made in different ways. Among them, one of the methods comprises the disease-causing virus passing via a series of embryos of animal (typically chick embryos) or cell culture [57,58]. Chick embryos can be taken as a host where the virus is grown in a series with different embryos [59]. In this way, the virus can diminish its strength to replicate in human cells whereas it increases its capability to replicate in host cells. A virus before using it as a vaccine, has to be grown in 200 different cell cultures or embryos. All these approaches involve transmitting a virus via a non-human host to create a new strain of the virus that can still be familiar to the immune system of humans, but in human host cells, they no longer replicate. When this vaccine is applied to humans it does not cause any illness, but still provides an immune response that can protect against future infection. During all these processes we should remember one thing in this vaccine virus can revert to a form that is capable of causing disease. Sometimes viruses mutate in such a way that may result in a virulent strain. So, during the development of an attenuated vaccine, one should consider all these facts. Rotavirus vaccine, MMR (measles, mumps, and rubella) vaccine, nasal flu vaccine, shingles vaccine, chickenpox vaccine (for specific groups only), BCG vaccine against TB (for special groups only), yellow fever vaccine, oral typhoid vaccine, etc are the examples of attenuated vaccine.
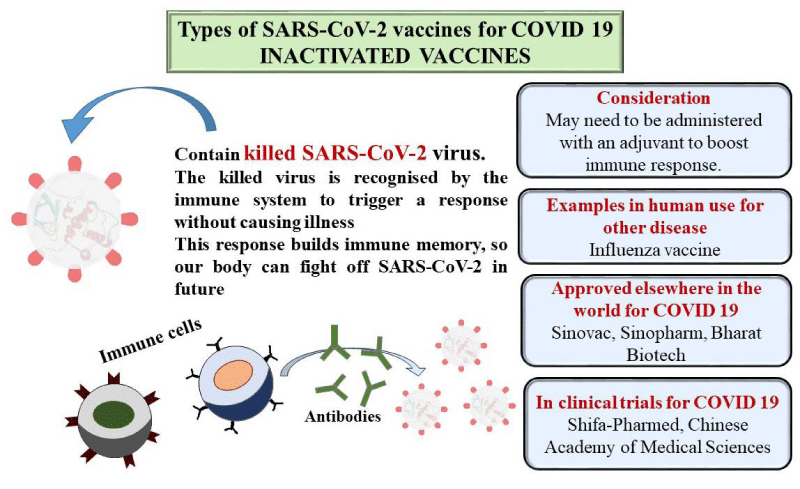
Killed or inactivated vaccines
Another type of vaccine is a killed or inactivated vaccine which is developed by inactivating a pathogen with the treatment of chemicals (formaldehyde or formalin) or using heat [60]. This treatment can destroy the replication capacity of the pathogen, although it can easily recognized by the immune system (the term “killed” is generally not used instead of “Inactivated” as this type of vaccine belongs to viral vaccines, and viruses are generally considered dead). Again killed or inactivated viruses can’t replicate at all; therefore they never return to a more virulent strain that may cause disease [61]. All these facts suggest that it provides short-term effects compared to live attenuated vaccines. Some examples of inactivated vaccines are flu vaccines, hepatitis A vaccines (special groups only), rabies vaccines, Japanese encephalitis vaccines, etc.
Recombinant vaccines
Generally, recombinant vaccines are prepared by using yeast or bacterial cells [62,63]. At first, a part of DNA is taken from bacteria or viruses from which we want to protect ourselves. Then make them active components in a large amount for the vaccine (generally with sugar or a single protein) by further inserting them into other cells. For example, the hepatitis B vaccine can be developed by taking a small part of the DNA from this vaccine and inserting it into the yeast cell DNA [64]. Therefore after purification of the vaccine, one of the surface proteins from the hepatitis B virus is used as an active component. Examples of these types of vaccines are the Hepatitis B vaccine, HPV vaccine, MenB vaccine, injected typhoid vaccine (a polysaccharide vaccine), etc.
mRNA vaccines
Vaccines can be developed by some intermediary between DNA and proteins, one such example is mRNA vaccines [65,66]. Some issues related to mRNA such as instability and their delivery into cells are overcome by recent technological advances. Apart from this some of the early results of mRNA vaccines are encouraging. A novel kind of RNA vaccine made up of the nucleic acid RNA which is available within one vector similar to lipid nanoparticles [67,68]. For example, Zika virus vaccines also different types of RNA vaccines are under progress to contest with the COVID-19 pandemic.
Recombinant vector vaccines
This type of vaccine involves delivery of genetic material into cells by using a harmless virus or bacterium as a vector, or carrier instead of delivering nucleic acid (DNA or mRNA) onto cells. Animals can be protected against infectious diseases such as rabies and distemper through numerous recombinant vector vaccines [69]. Examples of these types are HIV, Zika virus, and Ebola virus.
Mode of action of COVID-19 vaccines
Inactivated virus vaccines: This type of virus cannot reproduce within the body, so advanced doses are required [70]. Sometimes an adjuvant is used to strengthen the immune responses. This vaccine induces only antibody-mediated immunity. Since the microorganism is dead, the immune system can learn to recognize its antigen know-how to fight in the future as shown in Figure 3. Here, the immunological mode of action is started when neighboring cells are infected. These mechanisms of action are antigen-presentation and associated with CD4+ lymphocytes and SLA II and cytotoxicity activation related with CD8+ lymphocytes and SLA I, and then release of various cytokines. This type of vaccine was developed by the Sinovac company of China called CoronaVac while Covaxin was developed by Bharat Biotech and the ICMR jointly [71]. Both vaccines contain aluminum hydroxide; which acts as an adjuvant that boosts the efficiency of the vaccines [72,73]. In Covaxin, a supplementary adjuvant was used known as a Toll-Like Receptor (TLR) against the origin of dependable, which also prompts a strong immune response [74].
Figure 3: Mechanism of action of inactivated vaccine.
Inactivated vaccines are created by rendering a virulent virus noninfectious while maintaining its ability to stimulate an immune response; although safe, they require larger doses and multiple injections, along with booster shots over the years, to achieve levels of immunity similar to those provided by smaller doses of live attenuated vaccines which shown in Figure 3.
Viral subunit vaccines: Figure 4 represents schematically the mechanistic pathway of the viral protein subunit vaccine. Here virus antigen was used instead of genetic material; generally, an adjuvant is used to provide an elevated immune response [75,76]. This vaccine usually induces antibody-mediated immunity and with the aid of antigen-presenting cells, the antigens are known by T helper cells as a genuine viral disease [77,78]. For SARS-CoV-2 there are two subunit vaccines in phase 2 viz. the entire spike protein (Sanofi+GSK) or the receptor binding domain [79]. There are two subunits in the S Protein viz. S1 and S2 subunits. The S1 subunit consists of the following domains:
Figure 4: Mechanism of action of viral protein subunit.
(a) N-terminal domain (NTD)
(b) Receptor-binding domain (RBD)
(c) Receptor-binding motif (RBM) domains while the S2 subunit is made up of fusion peptide, heptad repeat 1 & 2. The entry of the virus into the cell through endocytosis by using the S-Protein interceded binding to the receptor of the hACE2. So, the S-protein along with its antigenic fragments is the key target of this vaccine [80]. The S glycoprotein has two conformational states viz. pre and post-fusion state. As a result, the antigen must retain its surface chemistry and shape of the new pre-fusion spike protein to save the epitomes for igniting superior-quality antibody responses [81]. Moreover, the target of the covered RBM since an antigen will improve the neutralizing antibody reaction and increase the overall efficiency of the vaccine.
The Novavax COVID-19 vaccine generally contains a protein or a polysaccharide - a sugar molecule or a blending of the two from a pathogen. In the Novavax vaccine the SARS-CoV-2 components are produced in-vitro before isolating directly from the virus [82].
Protein subunit vaccines, developed through recombinant methods or purification after growing the pathogen, use a specific subunit to activate the immune system and require adjuvants and booster doses for effective immunogenic response, which helps prevent severe side effects but necessitates multiple administrations for lasting immunity which is shown in Figure 4.
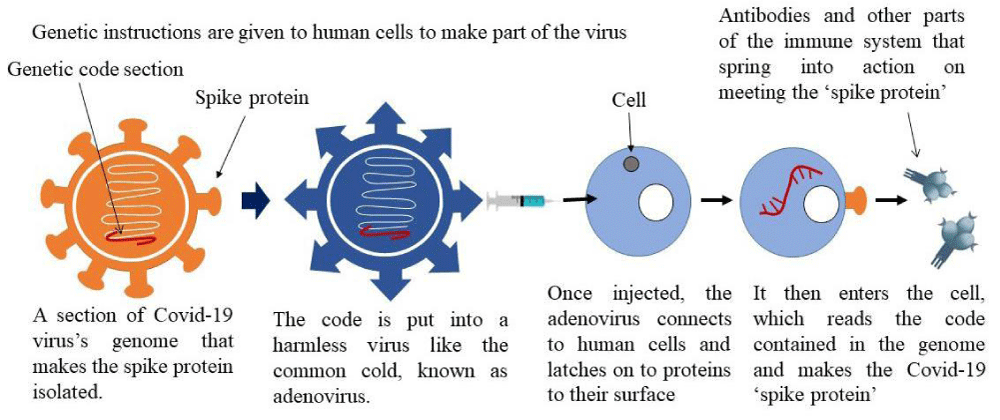
Viral vector vaccines: This vaccine may be of two types replicating and non-replicating. In non-replicating infecting a cell, creates the antigen of SARS-Cov-2 in that cell but not a novel virus whereas replication due to infection produces the antigen of SARS-CoV-2 in that cell and novel virus which affects other cells. The SARS-CoV-2 antigen can be seen within the cells since if the virus infects the body it produces cytotoxic T cells and helper cells [83]. The COVID-19 Oxford-AstraZeneca vaccine uses a chimpanzee universal freezing viral vector identified as ChAdOx1, which delivers the code that allows our cells to make the SARS-CoV-2 spike protein which is shown in Figure 5 [84].
Figure 5: Mechanistic pathway of adenovirus vaccine.
RNA vaccines
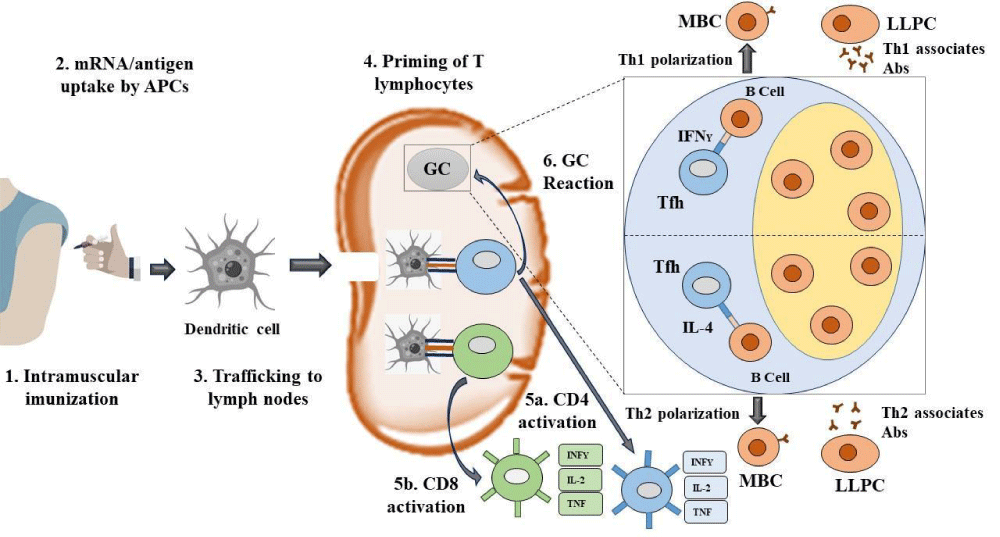
These vaccines transfer RNA-coding viral proteins into human cells. Then, they will replicate themselves and produce viral proteins through the aid of polymerase, and this way, RNA-based vaccines copy ongoing viral infection including toll-like receptors activation and IFN production [85,86]. Such vaccine versions of mRNA for the spike protein have been developed by Moderna, Curevac, and Pfizer [87,88]. RNA-based vaccines are relatively economical and easy to handle in case of mass production. The antigens of mRNA-LNPs are picked up by APCs viz. dendritic cells (DCs) [89]. Then these APCs enter the nodes of lymph where they are capable of keying CD4+ and CD8+ T lymphocytes [90]. The CD8+ T cells can initiate the creation of cytotoxic T lymphocytes (5b) which are able to homicide the infected cells. The CD4+ T cells can break up into two cells viz. Th1 cells or T follicular helper (Tfh) cells [91]. Tfh cells aid in beginning a germinal center (GC) reaction. GC reactions introduced by vaccination will affect the formation of antibody-secreting long-lived plasma cells (LLPCs) and memory B cells (MBCs). Tfh cells can be directed towards the phenotypes of Th1 or Th2, which will control the class switching of antibodies (Abs) formed by LLPCs to Th1- or Th2-related Absas shown in Figure 6 [92].
Figure 6: Mechanism of action of mRNA vaccine.
mRNA vaccines introduce a piece of genetic material that directs the body’s cells to create a viral protein, prompting the immune system to recognize it as foreign and generate antibodies to combat the actual virus in the future which is exhibited in Figure 6.
Current status of COVID-19 vaccine
To prevent the ongoing pandemic caused by coronavirus, a race started among scientists to create COVID-19 vaccines safely and effectively in a record time. Research and testing are required before vaccines can be used in clinics. To date, 97 COVID-19 vaccines are in clinical trials on humans and among them, 32 have reached the final stages (stage 3) of testing [93,94]. Currently, 10 vaccines are in early or emergency use whereas only 8 vaccines are approved for full use in several countries. Also, there are 5 vaccines that get abandoned after clinical trials. In Table 1, we have summarised the current status of some leading COVID-19 vaccines [94].
| Table 1: Current status of COVID -19 vaccine. | |||||
| Types | Developer | Dose | Phase | Status | Efficacy |
| mRNA | Pfizer-BioNTech | 2 doses, 3 weeks apart | 2, 3 | Approved in several countries, emergency use in the U.S., E.U., and other countries. | 91.3% |
| Moderna | 2 doses, 4 weeks apart | 3 | Approved in Switzerland, emergency use in the U.S., E.U., and other countries. | >90% | |
| Adenovirus | Oxford-Astrazeneca | 2 doses, 4 weeks apart | 2,3 | Approved in Brazil, emergency use in U.K, E.U., India, and other countries. | 76% |
| Gamaleya | 2 doses, 3 weeks apart | 3 | Emergency use in Russia, and other countries. | 91.6% | |
| Johnson & Johnson | Single dose | 3 | Emergency use in the U.S., E.U., and other countries. | 64-72% | |
| CanSino | Single dose | 3 | Approved in China, emergency use in other countries. | 65.28% | |
| Inactivated | Sinopharm | 2 doses, 3 weeks apart | 3 | Approved in China, U.A.E, Bahrain, and emergency use in other countries. | 78.1% |
| Sinovac | 2 doses, 2 weeks apart | 3 | Approved in China, emergency use in other countries. | 50.65% in Brazil, 83.5% in Turkey | |
| Bharat Biotech | 2 doses, 4 weeks apart | 3 | Emergency use in India, and other countries. | 77.8% | |
| Protein | Vector institute | 2 doses, 3 weeks apart | 3 | Early use in Russia, approved in Turkmenistan | Unknown |
| Novavax | 2 doses, 3 weeks apart | 3 | _ | 89.7% | |
| Centro De Ingenieria Genetica & Biotecnologia | 3 doses | 3 | Emergency use in Cuba | 92.28% | |
| DNA | Zydus | 3 doses, 4 weeks apart | 3 | _ | 66.6% |
Different variants of mutant COVID-19
Generally, viruses constantly change their characteristics through mutation [95,96]. Therefore a virus undergone one or more mutations is called a variant of the original virus. Since the emerging SARS-CoV-2 virus several variants that cause COVID-19 diseases by circulating around the world throughout the ongoing COVID-19 pandemic [97]. Most of the new variants have slightly different viral characteristics but few of them may possess reinforced viral properties such as spread rate, increase in disease severity, immunity to vaccine, or other collateral health issues.
According to WHO (World Health Organization), all the variants of the SARS-CoV-2 virus are classified as Variants of Concern (VOC) and Variants of Interest (VOI) [98,99]. Tables 2-4 represent the current status of VOC and VOI respectively.
| Table 2: Current status of Variants of Concern (VOC). | |||||
| Variant names | Pangolineages | Next strain clade | Earliest documented sample | Additional amino acid changes | Date of designation |
| Alpha | B.1.1.7 | 20I (V1) | U.K, Sept-2020 | +S:484K +S:452R |
18-12-2020 |
| Beta | B.1.351 B.1.351.2 B.1.351.3 |
20 H (V2) | South Africa, May-2020 | +S:L18F | 18-12-2020 |
| Gamma | P.1 P.1.1 P.1.2 |
20J(V3) | Brazil, Nov-2020 | +S:681H | 11-01-2021 |
| Delta | B.1.617.2 AY.1 AY.2 |
21A | India, Oct-2020 | +S:417N | VOI:04-04-2021 VOC:11-05-2021 |
| Table 3: Current status of Variants of Interest (VOI). | ||||
| Variant names | Pango lineages | Earliest documented sample | Next strain Clade | Date of designation |
| Eta | B.1.525 | Multiple countries, Dec-2020 | 21D | 17-03-2021 |
| Lota | B.1.526 | U.S.A, Nov-2020 | 21F | 24-03-2021 |
| Kappa | B.1.617.1 | India, Oct-2020 | 21B | 04-04-2021 |
| Lambda | C.37 | Peru, Dec-2020 | 21G | 14-06-2021 |
| Table 4: Mechanism and adverse effects of SARS-CoV-2 vaccines. | ||||
| COVID-19 vaccines | Possible mechanism | Vaccine components | Adverse effects | References |
| BNT16b2 | IgE-mediated reactions and non-IgE mediated reactions |
PEG, DSPC, trometamol | Anaphylaxis | [100,101] |
| Ad26.CoV2.S | Cross-reactivity with PEG | Polysorbate 80, | Anaphylaxis | [102] |
| mRNA-1273 | Spike binds to ACE2 receptor in cardiomyocytes | mRNA, spike protein, impurities | Myocarditis | [103,104] |
| Ad26.CoV2.S | Herpes zoster | Adenovirus, mRNA | trauma and immunosuppression | [105,106] |
| AZD1222 | Vaccine-induced immune thrombotic thrombocytopenia | Adenovirus, spike protein, EDTA, tPA, CAR receptor, sialic acid receptor, TLR ligands, impurities |
Thrombosis and thrombocytopenia |
[107,108] |
Challenges and future perspective of the COVID-19 vaccine
To take control of the pandemic situation, herd immunity is required by the immunization of a critical mass of the world population at once but data shows there are five risk factors that may delay the process of herd immunity. First, vaccination of people may prove lower than expected. That could happen if a real or perceived safety issue increases hesitancy or if younger people see little reason to be vaccinated once older groups are protected and a transition toward normalcy is well underway. Secondly, herd immunity relies on the efficacy of vaccines to reduce transmission, though the efficacy rate may not prove high enough to drive herd immunity. Third, the duration of immunity adopted by vaccines may prove shorter than expected which makes it hard to achieve simultaneous immunity. Fourth, supply-chain disruptions and delays may occur and this will produce supply shocks which will come in conflict with timelines. Fifth and most importantly, variants that reduce the efficacy of vaccines or the benefits of natural immunity may spread widely. Initial data indicates some concerning evidence that the examples of such variants are B.1.351 and P.1. Although the recent Novavax data shows that the vaccine is effective against severe diseases caused by B.1.351. Similarly, limited data from the Pfizer-BioNTech and AstraZeneca vaccines shows evidence of some protection against P.1. These five factors combined mean that there’s still an opportunity that herd immunity will not be achieved in the near future. Following the two endpoints, a transition toward normalcy and herd immunity may appear different in numerous geographical places. As soon as the new cases drop down may include a series of steps that will gradually normalize aspects of social and economic life though that will vary by geography. The much-awaited steps include a return of fully in-classroom education, reopening of restaurants and bars, gathering with larger groups of people, reopening of offices, etc. Not everyone will immediately resume all of their activities rather there will be a gradual or step-by-step shift toward more of them.
From various research works done by scientists and doctors, it may be concluded that ‘herd immunity’ is the only way to reach a more definitive end to the pandemic. Yet the most concerning fact is that new cases and deaths still indicate the virus may continue to circulate for one or more quarters after herd immunity is reached. Even when a nation reaches herd immunity, ongoing surveillance, booster vaccines, and potentially other measures may be needed. After almost 2 years later, the end of the pandemic is in sight for some parts of the world. It’s much too early to declare victory; however, we hope that the leaders and policymakers set much-needed policies and strategies to bring back the pre pandemic stage.
The worldwide SARS-CoV-2 pandemic has killed thousands, disrupted livelihoods, and caused widespread property and socioeconomic damage. It is crucial to understand the pathophysiology of SARS-CoV-2 infection and how it affects the immune system of the host cell in order to treat it. Currently, there is no antiviral medication available to combat SARS-CoV-2, therefore vaccination is the only way to safeguard against this viral attack, particularly for those who are at high risk. A variety of vaccines are available, including live attenuated vaccines, killed or inactivated vaccines, recombinant vaccines, mRNA vaccines, recombinant vector vaccines, and DNA vaccines. Pfizer-BioNTech and AstraZeneca vaccines show some protection against P.1, according to Novavax data. AstraZeneca vaccines reveal some protection against mild diseases caused by B.1.351. An attached protein called N (nucleocapsid) is located on the SARS-CoV-2 envelope membrane protein, S (spike), E (envelope), and M (membrane). A SARS-CoV-2 protein termed N regulates IFN- -synthesis and signaling by inhibiting IFN1 as it does in other coronaviruses. In contrast, the effectiveness of the innate immune system in fighting viral infections is mainly determined by IFN1 production and downstream signaling, which results in suppressing viral replication and triggering an effective adaptive response. An immunization of a critical mass of the world population simultaneously is necessary for herd immunity in the event of a pandemic. Therefore, the SARS-CoV-2 pandemic has led to the rapid development of vaccine formulations that could help us to strengthen our immune system in the event of future pandemics.
Declaration of competing interest
There are no known competing financial interests or personal relationships that could have appeared to influence the work reported in this review by the authors.
Ethics approval and consent to participate: This Study contains no animal or human studies. Hence ethical approval is not required.
Written consent for publication: All the authors consent for publication.
Availability of data and material: The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions: Swapan Kumar Chowdhury- Conceptualization, Writing. Niranjan Kumar Mridha- Formal analysis; Abdul Ashik Khan- Prepared tables; Nabajyoti Baildya- Data curation; Manab Mandal- Review and editing, Supervision; Narendra Nath Ghosh- Supervision.
- Chaplin DD. Overview of the immune response. J Allergy Clin Immunol. 2010;125:S3–23. Available from: https://www.jacionline.org/article/S0091-6749(09)02300-7/fulltext
- Møller AP, Saino N. Immune response and survival. Oikos. 2004;104:299–304. Available from: https://www.jstor.org/stable/3547960
- Parkin J, Cohen B. An overview of the immune system. Lancet. 2001;357:1777–89. Available from: https://www.sciencedirect.com/science/article/pii/S0140673600049047
- Medina KL. Overview of the immune system. Handb Clin Neurol. 2016;133:61–76. Available from: https://www.sciencedirect.com/science/article/pii/B9780444634320000049
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. The shape and structure of proteins. In: Molecular Biology of the Cell. 4th ed. New York: Garland Science; 2002. Available from: https://www.ncbi.nlm.nih.gov/books/NBK26830/
- Aristizábal B, González Á. Innate immune system. In: Autoimmunity: From Bench to Bedside [Internet]. Bogotá: El Rosario University Press; 2013. Available from: https://www.ncbi.nlm.nih.gov/books/NBK459455/
- Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782–7. Available from: https://www.nature.com/articles/35021228
- Netea MG, Schlitzer A, Placek K, Joosten LA, Schultze JL. Innate and adaptive immune memory: an evolutionary continuum in the host’s response to pathogens. Cell Host Microbe. 2019;25:13–26. Available from: https://www.sciencedirect.com/science/article/pii/S1931312818306273
- Roberts K, Alberts B, Johnson A, Walter P, Hunt T. Molecular biology of the cell. New York: Garland Science; 2002.
- Stockinger B, Zal T, Zal A, Gray D. B cells solicit their own help from T cells. J Exp Med. 1996;183:891–9. Available from: https://rupress.org/jem/article/183/3/891/47712/B-cells-solicit-their-own-help-from-T-cells
- Selders GS, Fetz AE, Radic MZ, Bowlin GL. An overview of the role of neutrophils in innate immunity, inflammation and host-biomaterial integration. Regen Biomater. 2017;4:55–68. Available from: https://academic.oup.com/rb/article/4/1/55/2799181
- Elkington P, O'Kane C, Friedland J. The paradox of matrix metalloproteinases in infectious disease. Clin Exp Immunol. 2005;142:12–20. Available from: https://onlinelibrary.wiley.com/doi/full/10.1111/j.1365-2249.2005.02840.x
- Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240–73. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2668232/
- Rose NR, Mackay IR. The autoimmune diseases. Amsterdam: Elsevier; 2006. Available from: https://pure.johnshopkins.edu/en/publications/the-autoimmune-diseases-10
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. Available from: https://www.sciencedirect.com/science/article/pii/S0092867406002340
- Amarante-Mendes GP, Adjemian S, Branco LM, Zanetti LC, Weinlich R, Bortoluci KR. Pattern recognition receptors and the host cell death molecular machinery. Front Immunol. 2018;9:2379. Available from: https://www.frontiersin.org/articles/10.3389/fimmu.2018.02379/full
- Jensen S, Thomsen AR. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol. 2012;86:2900–10. Available from: https://journals.asm.org/doi/10.1128/JVI.05738-11
- Schlee M, Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol. 2016;16:566–80. Available from: https://www.nature.com/articles/nri.2016.78
- Uehata T, Takeuchi O. RNA recognition and immunity—innate immune sensing and its posttranscriptional regulation mechanisms. Cells. 2020;9:1701. Available from: https://www.mdpi.com/2073-4409/9/7/1701
- Liu T, Zhang L, Joo D, Sun SC. NF-κB signaling in inflammation. Signal Transduct Target Ther. 2017;2:1–9. Available from: https://www.nature.com/articles/sigtrans201723
- Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. Available from: https://www.nature.com/articles/nri3581
- Feng H, Zhang YB, Gui JF, Lemon SM, Yamane D. Interferon regulatory factor 1 (IRF1) and anti-pathogen innate immune responses. PLoS Pathog. 2021;17:e1009220. Available from: https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1009220
- Ali S, Mann-Nüttel R, Schulze A, Richter L, Alferink J, Scheu S. Sources of Type I interferons in infectious immunity: plasmacytoid dendritic cells not always in the driver's seat. Front Immunol. 2019;10:778. Available from: https://www.frontiersin.org/articles/10.3389/fimmu.2019.00778/full
- Platanias LC. Mechanisms of type-I-and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–86. Available from: https://www.nature.com/articles/nri1604
- De Weerd NA, Nguyen T. The interferons and their receptors—distribution and regulation. Immunol Cell Biol. 2012;90:483–91. Available from: https://www.nature.com/articles/icb20129
- Nan Y, Wu C, Zhang YJ. Interplay between Janus kinase/signal transducer and activator of transcription signaling activated by type I interferons and viral antagonism. Front Immunol. 2017;8:1758. Available from: https://www.frontiersin.org/articles/10.3389/fimmu.2017.01758/full
- Pugnale P, Pazienza V, Guilloux K, Negro F. Hepatitis delta virus inhibits alpha interferon signaling. Hepatology. 2009;49:398–406. Available from: https://aasldpubs.onlinelibrary.wiley.com/doi/full/10.1002/hep.22654
- Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–45. Available from: https://www.annualreviews.org/doi/abs/10.1146/annurev-immunol-032713-120231
- Crotta S, Davidson S, Mahlakoiv T, Desmet CJ, Buckwalter MR, Albert ML, et al. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 2013;9:e1003773. Available from: https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1003773
- Rosa BA, Ahmed M, Singh DK, Choreño-Parra JA, Cole J, Jiménez-Álvarez LA, et al. IFN signaling and neutrophil degranulation transcriptional signatures are induced during SARS-CoV-2 infection. Commun Biol. 2021;4:1–14. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7418717/
- Sa Ribero M, Jouvenet N, Dreux M, Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16:e1008737. Available from: https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1008737
- Taefehshokr N, Taefehshokr S, Hemmat N, Heit B. Covid-19: Perspectives on innate immune evasion. Front Immunol. 2020;11:580641. Available from: https://www.frontiersin.org/articles/10.3389/fimmu.2020.580641/full
- Matsumiya T, Stafforini DM. Function and regulation of retinoic acid-inducible gene-I. Crit Rev Immunol. 2010;30(6):489–513. Available from: https://www.ingentaconnect.com/content/ben/cri/2010/00000030/00000006/art00010
- Sui L, Zhao Y, Wang W, Wu P, Wang Z, Yu Y, et al. SARS-CoV-2 membrane protein inhibits type I interferon production through ubiquitin-mediated degradation of TBK1. Front Immunol. 2021;12:662989. Available from: https://www.frontiersin.org/articles/10.3389/fimmu.2021.662989/full
- Seif F, Khoshmirsafa M, Aazami H, Mohsenzadegan M, Sedighi G, Bahar M. The role of JAK-STAT signaling pathway and its regulators in the fate of T helper cells. Cell Commun Signal. 2017;15:1–13. Available from: https://biosignaling.biomedcentral.com/articles/10.1186/s12964-017-0177-y
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. Available from: https://www.sciencedirect.com/science/article/pii/S0140673620301835
- Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. In: Seminars in immunopathology. Springer; 2017. p. 529–39. Available from: https://link.springer.com/article/10.1007/s00281-017-0629-x
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. T cells and MHC proteins. In: Molecular Biology of the Cell. 4th ed. New York: Garland Science; 2002. Available from: https://www.ncbi.nlm.nih.gov/books/NBK26926/
- Vazquez MI, Catalan-Dibene J, Zlotnik A. B cells responses and cytokine production are regulated by their immune microenvironment. Cytokine. 2015;74:318–26. Available from: https://www.sciencedirect.com/science/article/pii/S1043466615000786
- Crooke SN, Ovsyannikova IG, Kennedy RB, Poland GA. Immunoinformatic identification of B cell and T cell epitopes in the SARS-CoV-2 proteome. Sci Rep. 2020;10:1–15. Available from: https://www.nature.com/articles/s41598-020-70864-8
- Noorimotlagh Z, Karami C, Mirzaee SA, Kaffashian M, Mami S, Azizi M. Immune and bioinformatics identification of T cell and B cell epitopes in the protein structure of SARS-CoV-2: A systematic review. Int Immunopharmacol. 2020;106738. Available from: https://www.sciencedirect.com/science/article/pii/S1567576920302844
- Oliveira SC, de Magalhães MT, Homan EJ. Immunoinformatic analysis of SARS-CoV-2 nucleocapsid protein and identification of COVID-19 vaccine targets. Front Immunol. 2020;11:2758. Available from: https://www.frontiersin.org/articles/10.3389/fimmu.2020.01220/full
- Chang MS, Lu YT, Ho ST, Wu CC, Wei TY, Chen CJ, et al. Antibody detection of SARS-CoV spike and nucleocapsid protein. Biochem Biophys Res Commun. 2004;314:931–6. Available from: https://www.sciencedirect.com/science/article/pii/S0006291X03030464
- Chen X, Zhou B, Li M, Liang X, Wang H, Yang G, et al. Serology of severe acute respiratory syndrome: implications for surveillance and outcome. J Infect Dis. 2004;189:1158–63. Available from: https://academic.oup.com/jid/article/189/7/1158/813156
- Chen W, Xu Z, Mu J, Yang L, Gan H, Mu F, et al. Antibody response and viraemia during the course of severe acute respiratory syndrome (SARS)-associated coronavirus infection. J Med Microbiol. 2004;53:435–8. Available from: https://www.microbiologyresearch.org/content/journal/jmm/10.1099/jmm.0.45561-0
- Wang J, Wen J, Li J, Yin J, Zhu Q, Wang H, et al. Assessment of immunoreactive synthetic peptides from the structural proteins of severe acute respiratory syndrome coronavirus. Clin Chem. 2003;49:1989–96. Available from: https://academic.oup.com/clinchem/article/49/12/1989/5621101
- Li G, Chen X, Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N Engl J Med. 2003;349:508–9. Available from: https://www.nejm.org/doi/full/10.1056/NEJM200307313490520
- Kirkcaldy RD, King BA, Brooks JT. COVID-19 and postinfection immunity: limited evidence, many remaining questions. JAMA. 2020;323:2245–6. Available from: https://jamanetwork.com/journals/jama/fullarticle/2766097
- Fu Y, Pan Y, Li Z, Li Y. The utility of specific antibodies against SARS-CoV-2 in laboratory diagnosis. Front Microbiol. 2020;11:3312. Available from: https://www.frontiersin.org/articles/10.3389/fmicb.2020.603058/full
- Li CK, Wu H, Yan H, Ma S, Wang L, Zhang M, et al. T cell responses to whole SARS coronavirus in humans. J Immunol. 2008;181:5490–500. Available from: https://www.jimmunol.org/content/181/8/5490
- Shah VK, Firmal P, Alam A, Ganguly D, Chattopadhyay S. Overview of immune response during SARS-CoV-2 infection: lessons from the past. Front Immunol. 2020;11:1949. Available from: https://www.frontiersin.org/articles/10.3389/fimmu.2020.01949/full
- Seder RA, Ahmed R. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat Immunol. 2003;4:835–42. Available from: https://www.nature.com/articles/ni969
- Kleen TO, Galdon AA, MacDonald AS, Dalgleish AG. Mitigating coronavirus induced dysfunctional immunity for at-risk populations in COVID-19: trained immunity, BCG and “new old friends”. Front Immunol. 2020;11:2059. Available from: https://www.frontiersin.org/articles/10.3389/fimmu.2020.02059/full
- Buonaguro L, Pulendran B. Immunogenomics and systems biology of vaccines. Immunol Rev. 2011;239:197–208. Available from: https://onlinelibrary.wiley.com/doi/10.1111/j.1600-065X.2010.00971.x
- Drexler M. What you need to know about infectious disease. 2014. Available from: https://www.ncbi.nlm.nih.gov/books/NBK209710/
- Greenwood B. The contribution of vaccination to global health: past, present and future. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130433. Available from: https://royalsocietypublishing.org/doi/10.1098/rstb.2013.0433
- Li G, Gao X, Xiao Y, Liu S, Peng S, Li X, et al. Development of a live attenuated vaccine candidate against duck Tembusu viral disease. Virology. 2014;450:233–42. Available from: https://www.sciencedirect.com/science/article/pii/S0042682213007730
- Dai X, Xiong Y, Li N, Jian C. Vaccine types. In: Vaccines - the History and Future. IntechOpen; 2019. Available from: https://www.intechopen.com/chapters/65813
- Fenner F, Bachmann PA, Gibbs EPJ, Murphy FA, Studdert MJ, White DO. Cultivation and assay of viruses. Vet Virol. 1987;39. Available from: https://www.sciencedirect.com/science/article/pii/B9780122530555500074
- Sanders B, Koldijk M, Schuitemaker H. Inactivated viral vaccines. In: Vaccine analysis: strategies, principles, and control. Springer; 2015. p. 45–80. Available from: https://link.springer.com/chapter/10.1007/978-3-662-45024-6_2
- Burrell CJ, Howard CR, Murphy FA. Fenner and White's medical virology. Academic Press; 2016. Available from: https://www.sciencedirect.com/book/9780123751560/fenner-and-whites-medical-virology
- Soler E, Houdebine LM. Preparation of recombinant vaccines. Biotechnol Annu Rev. 2007;13:65–94. Available from: https://www.sciencedirect.com/science/article/abs/pii/S1387265607130040
- Andersson C. Production and delivery of recombinant subunit vaccines. Bioteknologi. 2000. Available from: https://www.diva-portal.org/smash/get/diva2:8775/FULLTEXT01.pdf
- Shouval D. Hepatitis B vaccines. J Hepatol. 2003;39:70–6. Available from: https://www.journal-of-hepatology.eu/article/S0168-8278(03)00152-1/fulltext
- Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines—a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–79. Available from: https://www.nature.com/articles/nrd.2017.243
- Xu S, Yang K, Li R, Zhang L. mRNA vaccine era—Mechanisms, drug platform and clinical prospection. Int J Mol Sci. 2020;21:6582. Available from: https://www.mdpi.com/1422-0067/21/18/6582
- Reichmuth AM, Oberli MA, Jaklenec A, Langer R, Blankschtein D. mRNA vaccine delivery using lipid nanoparticles. Ther Deliv. 2016;7:319–34. Available from: https://www.future-science.com/doi/10.4155/tde-2016-0006
- Aldosari BN, Alfagih IM, Almurshedi AS. Lipid nanoparticles as delivery systems for RNA-based vaccines. Pharmaceutics. 2021;13:206. Available from: https://www.mdpi.com/1999-4923/13/2/206
- da Fontoura Budaszewski R, Hudacek A, Sawatsky B, Krämer B, Yin X, Schnell MJ, et al. Inactivated recombinant rabies viruses displaying canine distemper virus glycoproteins induce protective immunity against both pathogens. J Virol. 2017;91:e02077-16. Available from: https://journals.asm.org/doi/full/10.1128/JVI.02077-16
- Pollard AJ, Bijker EM. A guide to vaccinology: from basic principles to new developments. Nat Rev Immunol. 2021;21:83–100. Available from: https://www.nature.com/articles/s41577-020-00479-7
- Kashte S, Gulbake A, El-Amin SF 3rd, Gupta A. COVID-19 vaccines: rapid development, implications, challenges and future prospects. Hum Cell. 2021;34:711–33. Available from: https://link.springer.com/article/10.1007/s13577-021-00512-4
- Liang Z, Zhu H, Wang X, Jing B, Li Z, Xia X, et al. Adjuvants for coronavirus vaccines. Front Immunol. 2020;11:2896. Available from: https://www.frontiersin.org/articles/10.3389/fimmu.2020.589833/full
- Hotez PJ, Corry DB, Strych U, Bottazzi ME. COVID-19 vaccines: neutralizing antibodies and the alum advantage. Nat Rev Immunol. 2020;20:399–400. Available from: https://www.nature.com/articles/s41577-020-0358-6
- Martínez-Flores D, Zepeda-Cervantes J, Cruz-Reséndiz A, Aguirre-Sampieri S, Sampieri A, Vaca L. SARS-CoV-2 vaccines based on the spike glycoprotein and implications of new viral variants. Front Immunol. 2021;12:701501. Available from: https://www.frontiersin.org/articles/10.3389/fimmu.2021.701501/full
- Coffman RL, Sher A, Seder RA. Vaccine adjuvants: putting innate immunity to work. Immunity. 2010;33:492–503. Available from: https://www.cell.com/immunity/fulltext/S1074-7613(10)00432-5
- Kyriakidis NC, López-Cortés A, González EV, Grimaldos AB, Prado EO. SARS-CoV-2 vaccines strategies: a comprehensive review of phase 3 candidates. NPJ Vaccines. 2021;6:1–17. Available from: https://www.nature.com/articles/s41541-021-00292-w
- Clem AS. Fundamentals of vaccine immunology. J Glob Infect Dis. 2011;3:73. Available from: https://doi.org/10.4103/0974-777x.77299
- Jeyanathan M, Afkhami S, Smaill F, Miller MS, Lichty BD, Xing Z. Immunological considerations for COVID-19 vaccine strategies. Nat Rev Immunol. 2020;20:615–32. Available from: https://www.nature.com/articles/s41577-020-00434-6
- Kaur SP, Gupta V. COVID-19 Vaccine: A comprehensive status report. Virus Res. 2020;198114. Available from: https://www.sciencedirect.com/science/article/pii/S0168170220304209
- Wang N, Shang J, Jiang S, Du L. Subunit vaccines against emerging pathogenic human coronaviruses. Front Microbiol. 2020;11:298. Available from: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00298/full
- Duan L, Zheng Q, Zhang H, Niu Y, Lou Y, Wang H. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: Implications for the design of spike-based vaccine immunogens. Front Immunol. 2020;11:2593. Available from: https://www.frontiersin.org/articles/10.3389/fimmu.2020.576622/full
- Tian JH, Patel N, Haupt R, Zhou H, Weston S, Hammond H, et al. SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat Commun. 2021;12:1–14. Available from: https://www.nature.com/articles/s41467-020-20653-8
- Costela-Ruiz VJ, Illescas-Montes R, Puerta-Puerta JM, Ruiz C, Melguizo-Rodríguez L. SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. Available from: https://www.sciencedirect.com/science/article/pii/S1359610120301036
- Mellet J, Pepper MS. A COVID-19 vaccine: big strides come with big challenges. Vaccines. 2021;9:39. Available from: https://www.mdpi.com/2076-393X/9/1/39
- Zhang C, Maruggi G, Shan H, Li J. Advances in mRNA vaccines for infectious diseases. Front Immunol. 2019;10:594. Available from: https://www.frontiersin.org/articles/10.3389/fimmu.2019.00594/full
- Said EA, Tremblay N, Al-Balushi MS, Al-Jabri AA, Lamarre D. Viruses seen by our cells: the role of viral RNA sensors. J Immunol Res. 2018;2018:9480497. Available from: https://www.hindawi.com/journals/jir/2018/9480497/
- Dolgin E. CureVac COVID vaccine let-down spotlights mRNA design challenges. Nature. 2021;594:483. Available from: https://www.nature.com/articles/d41586-021-01661-0
- Gaviria M, Kilic B. A network analysis of COVID-19 mRNA vaccine patents. Nat Biotechnol. 2021;39:546–8. Available from: https://www.nature.com/articles/s41587-021-00912-9
- Firdessa-Fite R, Creusot RJ. Nanoparticles versus dendritic cells as vehicles to deliver mRNA encoding multiple epitopes for immunotherapy. Mol Ther Methods Clin Dev. 2020;16:50–62. Available from: https://www.sciencedirect.com/science/article/pii/S2329050119301446
- Hirosue S, Dubrot J. Modes of antigen presentation by lymph node stromal cells and their immunological implications. Front Immunol. 2015;6:446. Available from: https://www.frontiersin.org/articles/10.3389/fimmu.2015.00446/full
- Luckheeram RV, Zhou R, Verma AD, Xia B. CD4+ T cells: differentiation and functions. Clin Dev Immunol. 2012;2012:925135. Available from: https://www.hindawi.com/journals/cdi/2012/925135/
- Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–42. Available from: https://www.cell.com/immunity/fulltext/S1074-7613(14)00441-9
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–27. Available from: https://www.nature.com/articles/s41586-020-2798-3
- Corum J, Grady D, Wee SL, Zimmer C. Coronavirus vaccine tracker. The New York Times. 2020;5. Available from: https://www.nytimes.com/interactive/2020/science/coronavirus-vaccine-tracker.html
- Lauring AS, Hodcroft EB. Genetic variants of SARS-CoV-2—what do they mean? JAMA. 2021;325:529–31. Available from: https://jamanetwork.com/journals/jama/fullarticle/2775006
- Khan AA, Dutta T, Mondal MP, Mandal SKC, Ahmed M, Baildya N, et al. Novel Coronavirus Disease (COVID-19): An extensive study on evolution, global health, drug targets and vaccines. Int J Clin Virol. 2021;5:54–69. Available from: https://www.heighpubs.org/jcv/jcv-aid1036.php
- Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int J Biol Sci. 2020;16:1678. Available from: https://www.ijbs.com/v16p1678.htm
- Konings F, Perkins MD, Kuhn JH, Pallen MJ, Alm EJ, Archer BN, et al. SARS-CoV-2 Variants of Interest and Concern naming scheme conducive for global discourse. Nat Microbiol. 2021;6:821–3. Available from: https://www.nature.com/articles/s41564-021-00932-w
- Resende PC, Naveca FG, Lins RD, Dezordi FZ, Ferraz MVF, Moreira EG, et al. The ongoing evolution of variants of concern and interest of SARS-CoV-2 in Brazil revealed by convergent indels in the amino (N)-terminal domain of the spike protein. Virus Evol. 2021;7(2):veab069. Available from: https://academic.oup.com/ve/article/7/2/veab069/6330639
- Quan PL, Ollé L, Sabaté-Brescó M, Guo Y, Muñoz-Cano R, Wagner A, et al. SARS-CoV-2 vaccine excipients polyethylene glycol and trometamol do not induce mast cell degranulation, in an in vitro model for non-IgE-mediated hypersensitivity. Front Allergy. 2022;3:1046545. Available from: https://www.frontiersin.org/articles/10.3389/falgy.2022.1046545/full
- Maurer J, Walles T, Wiese-Rischke C. Optimization of primary human bronchial epithelial 3D cell culture with donor-matched fibroblasts and comparison of two different culture media. Int J Mol Sci. 2023;24(4):4113. Available from: https://www.mdpi.com/1422-0067/24/4/4113
- Davidson RM. True or False? At Least 55 Undeclared Chemical Elements Have Been Detected by ICP-MS in COVID-19 “Vaccines”. Int J Vaccine Theory Pract Res. 2024;3(2):1394.1–1394.28.
- Davidson RM, Broudy D, Yanowitz S, Santiago D, Oller JW Jr. True or False? At Least 55 Undeclared Chemical Elements Have Been Detected by ICP-MS in COVID-19 “Vaccines”. Int J Vaccine Theory Pract Res. 2024;3(2):1394.1.
- Mondal P, Misra D, Chowdhury SK, Mandal V, Dutta T, Baildya N, et al. Exhaled volatile organic compounds (VOCs): A potential biomarkers for chronic disease diagnosis. Volatile Org Compd. 2021;50083. Available from: http://dx.doi.org/10.13140/RG.2.2.10135.50083
- Le Gars M, Hendriks J, Sadoff J, Ryser M, Struyf F, Douoguih M, et al. Immunogenicity and efficacy of Ad26.COV2.S: An adenoviral vector–based COVID‐19 vaccine. Immunol Rev. 2022;310(1):47–60. Available from: https://onlinelibrary.wiley.com/doi/10.1111/imr.13088
- Sun X, Yang Y, Meng X, Li J, Liu X, Liu H. PANoptosis: Mechanisms, biology, and role in disease. Immunol Rev. 2024;321(1):246–62. Available from: https://onlinelibrary.wiley.com/doi/10.1111/imr.13279
- Kircheis R. Coagulopathies after vaccination against SARS-CoV-2 may be derived from a combined effect of SARS-CoV-2 spike protein and adenovirus vector-triggered signaling pathways. Int J Mol Sci. 2021;22(19):10791. Available from: https://www.mdpi.com/1422-0067/22/19/10791
- Mandal M, Chowdhury SK, Khan AA, Baildya N, Dutta T, Misra D, et al. Inhibitory efficacy of RNA virus drugs against SARS-CoV-2 proteins: an extensive study. J Mol Struct. 2021;1234:130152. Available from: https://www.sciencedirect.com/science/article/pii/S0022286021004726