More Information
Submitted: February 21, 2024 | Approved: March 11, 2024 | Published: March 12, 2024
How to cite this article: Ju C, Zhou C, Deng Z, Gao J, Jiang W, et al. A Low-cost High-throughput Targeted Sequencing for the Accurate Detection of Respiratory Tract Pathogen. Int J Clin Virol. 2024; 8: 001-007.
DOI: 10.29328/journal.ijcv.1001056
Copyright License: © 2024 Ju C, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: RT-PCR; Targeted sequencing; SARS-CoV-2; Respiratory tract pathogen
A Low-cost High-throughput Targeted Sequencing for the Accurate Detection of Respiratory Tract Pathogen
Changyan Ju1#, Chengbosen Zhou2#, Zhezhi Deng3, Jingwei Gao4, Weizhao Jiang4, Hanbing Zeng4, Haiwei Huang3, Yongxiang Duan1 and David X Deng4*
1Nanshan Center for Disease Control and Prevention, Shenzhen, China
2Nanan Center for Disease Control and Prevention, Chongqing, China
3The First Affiliated Hospital of Sun Yut-sen University, China
4Guangdong Ardent BioMed Co., Ltd., China
#Contribute equally to the papers
*Address for Correspondence: David Deng,. Guangdong Ardent BioMed Co., Ltd. Email: [email protected]
Introduction: The current gold standard for SARS-CoV-2 diagnosis by real-time RT-PCR has limitations of gene numbers that can be detected. In this study, we developed a low-cost and high-throughput next-generation sequencing technology that can overcome the limitations of RT-PCR.
Methodology: A targeted sequencing panel (TSP) consisting of approximately 500 amplicons was designed that can simultaneously detect a broad range of gene loci of SARS-CoV-2 and genes for the most common viruses of respiratory infectious viruses in a single run of up to 96 samples. 448 samples and 31 control samples were examined independently with both TSP and RT-PCR, results were compared for accuracy and other indicators.
Results: TSP identified 50 SARS-CoV-2 positive samples with a 99.33% match to RT-PCR results. It is not surprising that TSP also identified multiple viral infections from 96 samples, whereas RT-PCR could not. TSP demonstrated its ability to conclude diagnosis for those undecided from RT-PCR tests.
Conclusion: Our data demonstrated that TSP is a fast and accurate test for detecting multiple pathogen infections of the respiratory tract.
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) outbreak, known as coronavirus disease 2019 (COVID-19), poses a massive threat to public health worldwide [1,2]. According to the World Health Organization (WHO), over 760 million cases and 6.9 million deaths have been recorded globally in the period from December 2019 to August 2023, and it is believed that the actual number could be higher. The coronavirus genome contains at least ten Open Reading Frames (ORFs), among which the first ORF (ORF1ab) constitutes about two-thirds of the viral RNA [3,4]. Four major structural proteins including spike protein (S), membrane protein (M), envelope protein (E), and nucleocapsid protein (N) [5,6] are encoded by the additional SARS-CoV-2 ORFs, which are found in one-third of the genome. The current gold standard for SARS-CoV-2 diagnosis is the detection of viral RNA via real-time RT-PCR assay with primers specifically designed to target a few genes, mostly ORFs and N genes [7,8]. The limitation of this technology may negatively affect detection rates and result in false negative reports [9]. For example, a number of deletions were observed in ORF8 and further deletion variants may emerge due to the immune-driven selection [10]. Phan, et al. performed a genetic analysis of 86 complete or near-complete SARS-CoV-2 genomes, revealing many mutations and deletions in coding and non-coding regions [11]. 116 mutations were identified in the ORF1ab gene from the research by Khailany, et al. [12]. It has been widely proven that a high mutation rate drives the evolution of virus and genome variation, thus enabling the virus to evade host immunity and develop drug resistance [13].
Detection of other common viral pathogens that typically cause seasonal respiratory tract infection has mainly been neglected in these years. Studies have shown that [14] there may be synergistic effects of coinfection compared to SARS-CoV-2 virus infection alone, resulting in a higher risk of death in patients. Patients with SARS-CoV-2 coinfected with other respiratory viruses are more likely to be admitted to ICU [15]. Most respiratory infections, however, are clinically symptomatic alike and indistinguishable between SARS-CoV-2 and other viruses, and current routine laboratory tests alone failed to distinguish SARS-CoV-2 from other respiratory viral infections [16]. Therefore, high-throughput and low-cost methods are in urgent need of precision diagnosis of respiratory viral infection. Here, we developed a low-cost, faster turnaround time and high-throughput Targeted Sequencing Panel (TSP) of approximately 500 amplicons that simultaneously detects a wide range of genetic loci for both SARS-CoV-2 and other most common respiratory viruses.
The Institutional Review Board (IRB) of the Nanshan Center for Disease Control and Prevention approved the study, and the IRB approval number for the research protocol is 2020A001.
Samples
This study was approved by the Institutional Review Board of the Nanshan Center for Disease Control and Prevention and conducted at Guangdong Ardent BioMed Co., Ltd. Written informed consent was obtained from all subjects. 448 nasopharyngeal swabs, 15 synthetic positive controls, and 16 Non-Template Controls (NTCs) were evaluated in this study. 419 samples were collected from the SARS-CoV-2 screen population in the Nanshan Center for Disease Control and Prevention, and 29 from the fever clinic of Guangzhou Eighth People’s Hospital. Nasopharyngeal swabs were immersed in 3 mL of Hank’s solution and transferred to the laboratory within 2 hours.
Design of TSP
Primers were designed based on the information from the database of 38 pathogens and 528 loci listed in Table 1. These 38 pathogens include the most prevalent respiratory viruses. Supplementary Data of detailed information is provided in Table S1.
| Table 1: Primers for targeted sequencing panel. | |||
| Pathogens | Types | Locus number | Gene or pathogens |
| SARS-CoV-2 | 1 | 66 | N, E, S of Orf1ab |
| Other coronaviruses | 7 | 23 | SARS-CoV, MERS-CoV, HCoV-229E, HCoV-HKU1, HCoV-NL63, HCoV-OC43 and EboV |
| Common respiratory RNA viruses | 12 | 48 | H9N2, H5N1, H3N2, H2N2, H7N9, H1N1, Flu-B, HPIV, HRSV-A, HRSV-B, HMPV-A and HMPV-B |
| Common respiratory DNA viruses | 16 | 389 | HAdV-5, HAdV-A, HAdV-B, HAdV-C, HAdV-D, HAdV-E, HHV-1, HHV-2, HHV-3, HHV-4, HHV-5, HHV-7, HPV-B19, JCPyV, SV40 and BKV |
| Candida glabrata and Candida parapsilosis | 2 | 2 | |
DNA/RNA Extraction and RT-PCR
The total DNA/RNA was extracted using the virus DNA/RNA extraction kit (Tianlong Technology, Xian, China). Briefly, samples were vortexed for 15 seconds, and 200 µL of each sample was used for DNA/RNA extraction. RT-PCR amplification was then performed on the PCR system (ABI 7500, Thermo Fisher Scientific, Waltham, MA, USA) using a commercial SARS-CoV-2 detection kit (Daan Gene, Guangzhou, China) following the instructions provided with the kit. The method targets specific genomic regions of SARS-CoV-2, orf1ab, and nucleocapsid (N) genes. Samples were considered positive if the Ct value of FAM and VIC channel was ≤ 37.0, or, the Ct value falls in between 37 and 40 for both duplicated tests.
Library preparation and sequencing
A DNA AmpliSeq library was established by multiplex RT-PCR. Briefly, 1 µL extracted DNA was mixed with 5 µL 2× primer panel, 2 µL AMPure XP containing 5× amplification enzyme mixture, Master Mix, and sequence beads, and 2 µL DNase-free water. PCR primers that are complementary to the adapters of the library will be linked to the surface of the beads. The target fragment amplification was carried out through PCR reaction with 1 cycle at 99 ℃ for 2 mins, 1 cycle at 99 ℃ for 15s, 20 cycles at 60 ℃ for 4 mins, followed by another cycle of synergy by adding 2 µL synergist reagent and continued with 50 ℃ for 10 mins, 55 ℃ for 10 mins, 60 ℃ for 20 mins, and 10 ℃ hold. The end product was mixed with 2 µL linker mixture, 1 µL DNA ligase, and 1 µL sequence linker, and perform linker ligation at 22 ℃ for 30 mins, 68 ℃ for 5 mins, 72 ℃ for 5 mins and held at 16 ℃ prior to purification. The final product was eluted with DNase-free water and mixed with 1 µL library enrichment primers, and 5 µL library enrichment enzyme mixture, to enrich the amplified cDNA strand. Purified magnetic beads were then added to the enriched library for the second round of purification, and 25 µL TE buffer was used for elution. The product was subjected to sequencing on an Ion torrent PGM sequencing system after qualification with Agilent 2100 bioanalyzer.
Dataset processing and quality control
Initial quality control was performed to retain the reads with double-ended lengths > 60 bp. To generate high-quality data, secondary quality control was performed to retain the reads with Q30 > 85%. Sequencing depth, uniformity, and on-target rate were also evaluated.
Bioinformatic analysis
Sequencing data were submitted to the Partek analysis pipeline for pathogen detection. Each virus’s pathogen detection results and genome coverage were obtained in base space sequence analysis. A sample is considered positive for SARS-CoV-2 with more than 3 distinct regions’ amplicons in this panel. The results of other common viruses are considered positive if there have been more than three reads found at any locus. Heatmap of the reads number of SARS-CoV-2 was generated using online bioinformatic tools.
(http://www.cloud.biomicroclass.com/CloudPlatform/SoftPage/Heatmap)
Determination of accuracy
Performance indicators of samples were calculated by comparing the results with RT-PCR. Five performance indicators, including Positive Percent Agreement (PPA), Negative Percent Agreement (NPA), accuracy, False Negative Rate (FNR), and False Positive Rate (FPR), were evaluated.
Determination of assay efficiency
To determine the Limit of Detection (LOD) of the SARS-CoV-2 virus, we used healthy human RNA mixed with SARS-CoV-2 and SARS-CoV virus standards (1 pg, 0.1 pg, 0.01 pg, and 0.001 pg), the titers of isolated virus standards were measured in SI derived unit (pg/mL). The sequencing process was conducted according to the NGS instruction above.
Limit of detection for SARS-CoV-2
Analytical sensitivity was assessed by determining the LOD using healthy human RNA containing SARS-CoV-2 and SARS-CoV standard. The SARS-CoV-2 standard (1, 0.1, 0.01, 0.001 pg) was mixed in 4 copies of human RNA as positive control and human RNA with water as negative control. The LOD was determined to be 0.01 pg/mL standard (Table 2).
| Table 2: Read numbers for the limit ofdetection of the SARS-CoV-2 standard. | ||||||
| Samples | SARS-CoV-2standard | SARS-CoVstandard | Result | |||
| ORF1ab | S | E | ||||
| human RNA | 0 | 0 | 0 | 0 | Negative | |
| human RNA+ 1 pg/mL Standard | 70118 | 0 | 0 | 95887 | Positive | |
| human RNA+ 0.1 pg/mL Standard | 5310 | 0 | 0 | 2341 | Positive | |
| human RNA+ 0.01 pg/mL Standard | 240 | 0 | 0 | 327 | Positive | |
| human RNA+ 0.001 pg/mL Standard | 1 | 0 | 0 | 0 | Negative | |
| water(blank) | 0 | 0 | 0 | 0 | Negative | |
Metric performance evaluation
All 448 samples were subjected to TSP on the Ion torrent platform independent of RT-PCR assay. One TSP run included 96 samples, and 11 batches of 448 samples were sequenced with fifteen positive controls and sixteen negative controls on each run. The overall accuracy of SARS-CoV-2 detection in comparison to RT-PCR is 99.33%. The PPA and NPA of the targeted sequencing were 96.08% and 99.75%, respectively (Table 3). The calculation formulas for PPA, NPA, FPR, FNR, and Accuracy are below.
Table 3: Sequencing results: concordance with RT-PCR. |
||||
| RT-PCR | ||||
| Positive | Negative | Total | ||
| SARS-CoV-2 targeted sequencing panel | Positive | 49 | 1 | 50 |
| Negative | 2 | 396 | 398 | |
| Total | 51 | 397 | 448 | |
PPA=TP/(TP+FN) =49/(49+2)=96.08%
NPA=TN/(TN+FP) =396/(396+1)=99.75%
Accuracy=TP+TN/All Results=(49+396)/448=99.33%
FPR=FP/(FP+TN) =1/(1+396)=0.25%
FNR=FN/(FN+TP) =2/(2+49)=3.92%
TSP of SARS-CoV-2
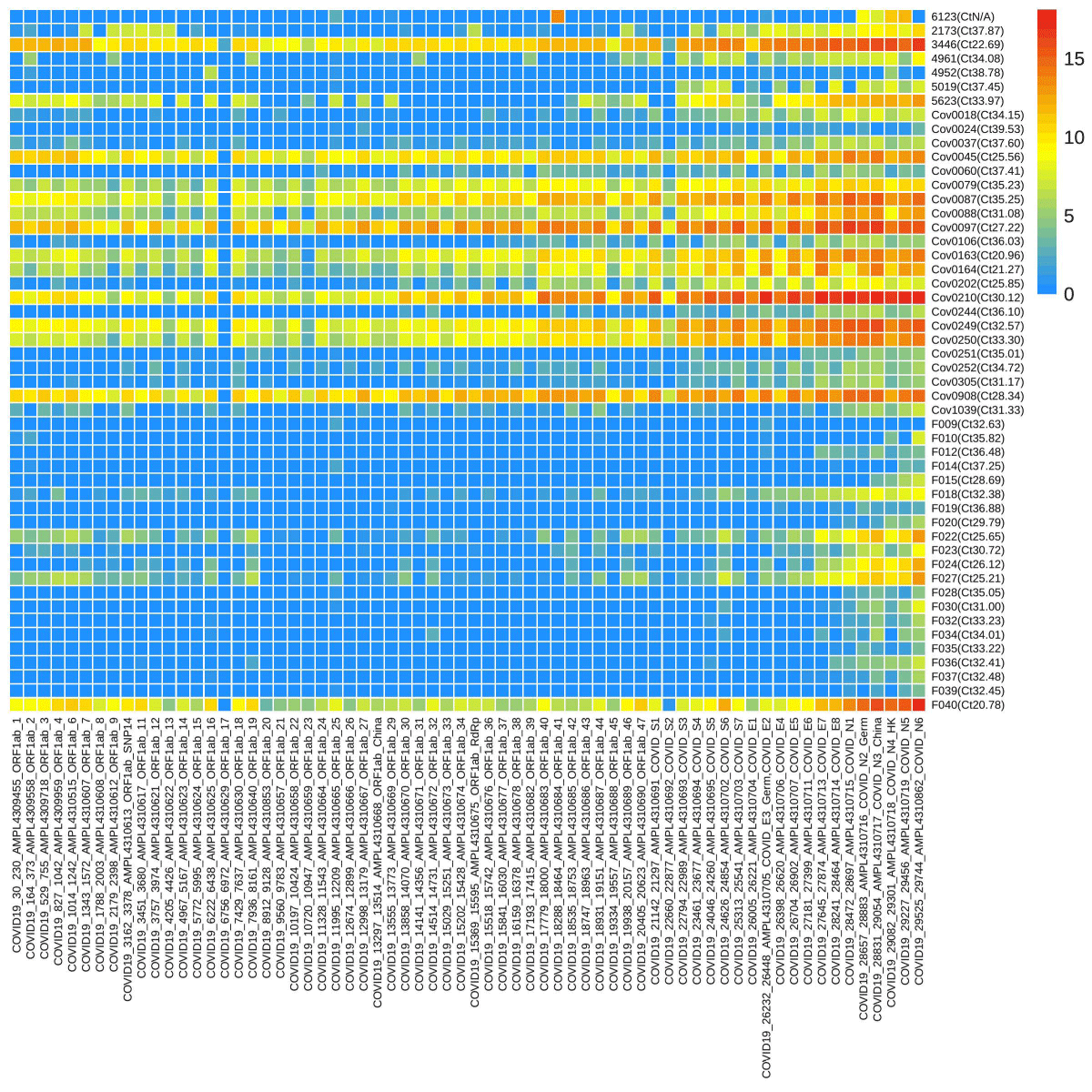
Sixty-six loci of SARS-CoV-2 were screened through targeted sequencing and 50 samples were found positive. The read numbers of various loci were significantly different ranging from 0 to 432369. A heatmap of SARS-CoV-2 was generated with a logarithmic value of reads number with a base of 2 from 50 positive samples (Figure 1). CoV0018-CoV1039 had more reads compared to F009-F039’s lower number. Full data were provided in Supplementary Data Table S2.. One sample was confirmed positive from TSP while having negative RT-PCR results. In addition, 4 samples exhibited undecided results in a single RT-PCR test, and 2 of them were confirmed positive through TSP.
Figure 1: The heatmap of reads numbers of SARS-CoV-2 by targeted sequencing. The labels on the Y-axis represent the names of samples, and the number in the bracket was the Ct value of the orf1ab gene by RT-PCR, while the X-axis represents the name of the loci detected. The various color denotes the reads number, a logarithmic value with a base of 2.
TSP of other respiratory pathogens
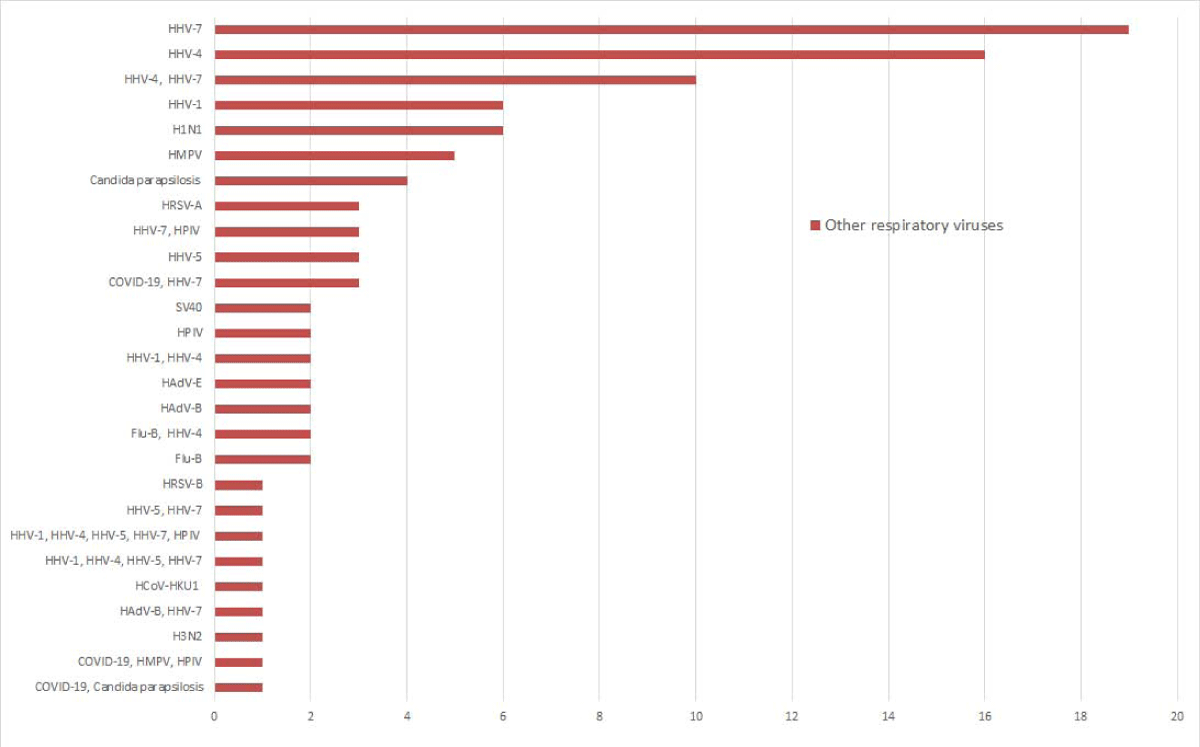
Of the 448 samples, n = 5 tested concurrent infections of SARS-CoV-2 with HHV-7, HMPV, HPIV, and Candida parapsilosis, n = 96 tested positive for at least one other viral pathogen, among which 21 were detected coinfection of at least two viral pathogens. Of the detected other respiratory viral pathogens, HHV (n = 68), HPIV (n = 7), and H1N1 (n = 6) were the most detected pathogens. 37.9% (11/29) of patients at the fever clinic had HHV infection, while 13.6% (57/419) of the SARS-CoV-2 screen population carried HHV. There was a statistical difference in the prevalence of HHV between the patient and the asymptomatic population (χ2 = 12.468, p < 0.05). Other viral pathogens detected were HMPV (n = 5), HAdV (n = 5), HRSV (n = 4), and Flu-B (n = 4), (Figure 2). Among the 5 samples with Candida parapsilosis infection, the reads number were 5, 10, 173, 431, and 110885. And one of them was coinfection with SARS-CoV-2.
Figure 2: Other respiratory viruses were detected by the targeted sequencing panel. The X-axis represents the number of positive samples, and the Y-axis represents the respiratory viruses detected by targeted sequencing.
The spread of 2019-nCoV worldwide brings huge pressure to public health [17,18]. RT-PCR has become the standard method for screening and diagnosis of the virus infection [19]. Nonetheless, while it demonstrated a huge advantage in large population screening [20,21], the limitation with targeted microorganisms hinders the use of this technology for the diagnosis of multiple co-infections. NGS technology has been used not only to demonstrate the origin of the novel coronavirus but also for virus detection, transmission, and mutation monitoring [22-24]. We used TSP in this study by combining specific biotinylated probes and hybrid capture enrichment that targeted 36 viruses and 2 fungi in one panel. The data demonstrated that TSP is able to identify multiple co-infections simultaneously, develop precision diagnosis of questionable samples from RT-PCR, and lower costs that might generated from repeated testing of RT-PCR.
The performance evaluation showed that our panel has a SARS-CoV-2 LOD of 0.01pg/mL, making the analysis highly sensitive and accurate for detecting the SARS-CoV-2 genome. The consistency between the TSP and RT-PCR was 99.33%. Performance evaluation showed that PPA, NPA, FPR, and FNR were 96.08%, 99.75%, 0.25%, and 3.92%, respectively. Moreover, the RT-PCR kit used in this study detects only one respiratory pathogen (SARS-CoV-2), and could not detect multiple viral infections.
Our sequencing results showed that one sample that was negative for ORF1ab on RT-PCR had a high copy fragment of the orf1ab gene, also helped to confirm another two samples that were questionable from RT-PCR, indicating that the advantage of TSP to assist RT-PCR in improving the true positive detection rate of the diagnosis.
NGS includes PCR amplicon sequencing, target enrichment sequencing, and metagenomic sequencing [25,26]. The advantage of targeted sequencing over the amplicon-based method is that it is based on many fragments and probes. It can also significantly reduce the sequencing depth, lower the detection cost, and with more accessible analytical performance compared to metagenomic sequencing [27-29].
There are no standard criteria for results interpretation for targeted sequencing. Thorburn, et al. [30] deemed samples with fewer than 10 unique viral reads to be negative by NGS. In Gaston’s study [28], raw read counts for viruses of ≥ 1 were taken forward for result interpretation. Based on our experience, a sample is considered positive for SARS-CoV-2 or other virus with more than 3 distinct regions’ amplicons in our panel. The results of other common viruses are considered positive if there have been more than three reads found at any locus.
This targeted sequencing relies on educated microbial prediction that may limit it from identifying unknown pathogens. Moreover, we detected only viruses but not bacteria in this study. The limited sample size may affect the representative geographically.
We developed a custom-designed TSP with high sensitivity and specificity for detecting SARS-CoV-2 and other respiratory viruses using Ion Torrent’s enrichment workflow, hybrid capture method, and bioinformatics pipeline. The low cost and high sample throughput analysis make it suitable for large-scale accurate detection of respiratory viruses.
Funding
This study was funded by the Key Discipline Construction Subsidy of Nanshan District, the Nanshan District Science (Infection disease prevention and control), and the Nanshan District Science and Technology Plan Project (2020006). The study design, data collection, interpretation, and the choice to submit the study for publication were all independent of the funders.
We appreciate Professor Maojun Zhang from the State Key Laboratory of Infectious Disease Prevention and Control, Chinese Center for Disease Control and Prevention, who helped with the manuscript writing.
- Malik YA. Properties of Coronavirus and SARS-CoV-2. Malays J Pathol. 2020 Apr;42(1):3-11. PMID: 32342926.
- Machkovech HM, Hahn AM, Garonzik Wang J, Grubaugh ND, Halfmann PJ, Johnson MC, Lemieux JE, O'Connor DH, Piantadosi A, Wei W, Friedrich TC. Persistent SARS-CoV-2 infection: significance and implications. Lancet Infect Dis. 2024 Feb 7:S1473-3099(23)00815-0. doi: 10.1016/S1473-3099(23)00815-0. Epub ahead of print. PMID: 38340735.
- Tam D, Lorenzo-Leal AC, Hernández LR, Bach H. Targeting SARS-CoV-2 Non-Structural Proteins. Int J Mol Sci. 2023 Aug 20;24(16):13002. doi: 10.3390/ijms241613002. PMID: 37629182; PMCID: PMC10455537.
- Hassanin AA, Haidar Abbas Raza S, Ahmed Ujjan J, Aysh ALrashidi A, Sitohy BM, Al-Surhanee AA, Saad AM, Mohamed Al-Hazani T, Osman Atallah O, Al Syaad KM, Ezzat Ahmed A, Swelum AA, El-Saadony MT, Sitohy MZ. Emergence, evolution, and vaccine production approaches of SARS-CoV-2 virus: Benefits of getting vaccinated and common questions. Saudi J Biol Sci. 2022 Apr;29(4):1981-1997. doi: 10.1016/j.sjbs.2021.12.020. Epub 2021 Dec 13. PMID: 34924802; PMCID: PMC8667566.
- Noori R, Sardar M. An outlook on potential protein targets of COVID-19 as a druggable site. Mol Biol Rep. 2022 Nov;49(11):10729-10748. doi: 10.1007/s11033-022-07724-3. Epub 2022 Jul 6. PMID: 35790657; PMCID: PMC9256362.
- Young M, Crook H, Scott J, Edison P. Covid-19: virology, variants, and vaccines. BMJ Med. 2022 Apr 1;1(1):e000040. doi: 10.1136/bmjmed-2021-000040. PMID: 36936563; PMCID: PMC9951271.
- Xu J, Chen J, Wen F, Liu K, Chen Y. Detection methods and dynamic characteristics of specific antibodies in patients with COVID-19: A review of the early literature. Heliyon. 2024 Jan 24;10(3):e24580. doi: 10.1016/j.heliyon.2024.e24580. PMID: 38317938; PMCID: PMC10839880.
- Caixeta DC, Paranhos LR, Blumenberg C, Garcia-Júnior MA, Guevara-Vega M, Taveira EB, Nunes MAC, Cunha TM, Jardim ACG, Flores-Mir C, Sabino-Silva R. Salivary SARS-CoV-2 RNA for diagnosis of COVID-19 patients: a systematic revisew and meta-analysis of diagnostic accuracy. Jpn Dent Sci Rev. 2023 Jun 21;59:219–38. doi: 10.1016/j.jdsr.2023.06.004. Epub ahead of print. PMID: 37360001; PMCID: PMC10284464.
- Kanji JN, Zelyas N, MacDonald C, Pabbaraju K, Khan MN, Prasad A, Hu J, Diggle M, Berenger BM, Tipples G. False negative rate of COVID-19 PCR testing: a discordant testing analysis. Virol J. 2021 Jan 9;18(1):13. doi: 10.1186/s12985-021-01489-0. PMID: 33422083; PMCID: PMC7794619.
- Su YCF, Anderson DE, Young BE, Linster M, Zhu F, Jayakumar J, Zhuang Y, Kalimuddin S, Low JGH, Tan CW, Chia WN, Mak TM, Octavia S, Chavatte JM, Lee RTC, Pada S, Tan SY, Sun L, Yan GZ, Maurer-Stroh S, Mendenhall IH, Leo YS, Lye DC, Wang LF, Smith GJD. Discovery and Genomic Characterization of a 382-Nucleotide Deletion in ORF7b and ORF8 during the Early Evolution of SARS-CoV-2. mBio. 2020 Jul 21;11(4):e01610-20. doi: 10.1128/mBio.01610-20. PMID: 32694143; PMCID: PMC7374062.
- Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol. 2020 Jul;81:104260. doi: 10.1016/j.meegid.2020.104260. Epub 2020 Feb 21. PMID: 32092483; PMCID: PMC7106203.
- Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020 Jun;19:100682. doi: 10.1016/j.genrep.2020.100682. Epub 2020 Apr 16. PMID: 32300673; PMCID: PMC7161481.
- Pachetti M, Marini B, Benedetti F, Giudici F, Mauro E, Storici P, Masciovecchio C, Angeletti S, Ciccozzi M, Gallo RC, Zella D, Ippodrino R. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020 Apr 22;18(1):179. doi: 10.1186/s12967-020-02344-6. PMID: 32321524; PMCID: PMC7174922.
- Stowe J, Tessier E, Zhao H, Guy R, Muller-Pebody B, Zambon M, Andrews N, Ramsay M, Lopez Bernal J. Interactions between SARS-CoV-2 and influenza, and the impact of coinfection on disease severity: a test-negative design. Int J Epidemiol. 2021 Aug 30;50(4):1124-1133. doi: 10.1093/ije/dyab081. PMID: 33942104; PMCID: PMC8135706.
- Garcia-Vidal C, Sanjuan G, Moreno-García E, Puerta-Alcalde P, Garcia-Pouton N, Chumbita M, Fernandez-Pittol M, Pitart C, Inciarte A, Bodro M, Morata L, Ambrosioni J, Grafia I, Meira F, Macaya I, Cardozo C, Casals C, Tellez A, Castro P, Marco F, García F, Mensa J, Martínez JA, Soriano A; COVID-19 Researchers Group. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: a retrospective cohort study. Clin Microbiol Infect. 2021 Jan;27(1):83-88. doi: 10.1016/j.cmi.2020.07.041. Epub 2020 Jul 31. PMID: 32745596; PMCID: PMC7836762.
- Singhal T. A Review of Coronavirus Disease-2019 (COVID-19). Indian J Pediatr. 2020 Apr;87(4):281-286. doi: 10.1007/s12098-020-03263-6. Epub 2020 Mar 13. PMID: 32166607; PMCID: PMC7090728.
- Herrera-Plasencia PM, Enoki-Miñano E, Ruiz-Barrueto MYA. Riesgos, contaminación y prevención frente al COVID-19 en el quehacer odontológico: una revisión [Risks, contamination and prevention against COVID-19 in dental work: a review]. Rev Salud Publica (Bogota). 2020 Sep 1;22(5):560-565. Spanish. doi: 10.15446/rsap.V22n5.86065. PMID: 36753227.
- Kaye AD, Okeagu CN, Pham AD, Silva RA, Hurley JJ, Arron BL, Sarfraz N, Lee HN, Ghali GE, Gamble JW, Liu H, Urman RD, Cornett EM. Economic impact of COVID-19 pandemic on healthcare facilities and systems: International perspectives. Best Pract Res Clin Anaesthesiol. 2021 Oct;35(3):293-306. doi: 10.1016/j.bpa.2020.11.009. Epub 2020 Nov 17. PMID: 34511220; PMCID: PMC7670225.
- Ma L, Zhu H, Jiang Y, Kong X, Gao P, Liu Y, Zhao M, Deng G, Cao Y. Development of a Novel Multiplex PCR Method for the Rapid Detection of SARS-CoV-2, Influenza A Virus, and Influenza B Virus. Int J Anal Chem. 2024 Feb 29;2024:4950391. doi: 10.1155/2024/4950391. PMID: 38456096; PMCID: PMC10919977.
- Samsunder N, Devnarain N, Sivro A, Kharsany ABM. The Performance of Diagnostic Tests for Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in the South African Population: A Scoping Review. Trop Med Infect Dis. 2023 Dec 1;8(12):514. doi: 10.3390/tropicalmed8120514. PMID: 38133446; PMCID: PMC10748306.
- Vasei M, Jafari E, Falah Azad V, Safavi M, Sotoudeh M. Molecular Diagnosis of COVID-19; Biosafety and Pre-analytical Recommendations. Iran J Pathol. 2023 Summer;18(3):244-256. doi: 10.30699/IJP.2023.1988405.3061. Epub 2023 Jul 16. PMID: 37942195; PMCID: PMC10628373.
- Carpenter RE, Tamrakar VK, Almas S, Sharma A, Sharma R. SARS-CoV-2 Next Generation Sequencing (NGS) data from clinical isolates from the East Texas Region of the United States. Data Brief. 2023 Aug;49:109312. doi: 10.1016/j.dib.2023.109312. Epub 2023 Jun 14. PMID: 37346925; PMCID: PMC10264325.
- Chan AP, Siddique A, Desplat Y, Choi Y, Ranganathan S, Choudhary KS, Khalid MF, Diaz J, Bezney J, DeAscanis D, George Z, Wong S, Selleck W, Bowers J, Zismann V, Reining L, Highlander S, Brown K, Armstrong JR, Hakak Y, Schork NJ. A CRISPR-enhanced metagenomic NGS test to improve pandemic preparedness. Cell Rep Methods. 2023 Apr 18;3(5):100463. doi: 10.1016/j.crmeth.2023.100463. PMID: 37323571; PMCID: PMC10110940.
- Tartanian AC, Mulroney N, Poselenzny K, Akroush M, Unger T, Helseth DL, Jr., Sabatini LM, Bouma M, Larkin PMK. NGS implementation for monitoring SARS-CoV-2 variants in Chicagoland: An institutional perspective, successes and challenges. Front Public Health. 2023;11:1177695. doi:10.3389/fpubh.2023.1177695 [Pubmed: 37151582]
- Wang X, Stelzer-Braid S, Scotch M, Rawlinson WD. Detection of respiratory viruses directly from clinical samples using next-generation sequencing: A literature review of recent advances and potential for routine clinical use. Rev Med Virol. 2022 Sep;32(5):e2375. doi: 10.1002/rmv.2375. Epub 2022 Jul 1. PMID: 35775736; PMCID: PMC9539958.
- Nafea AM, Wang Y, Wang D, Salama AM, Aziz MA, Xu S, Tong Y. Application of next-generation sequencing to identify different pathogens. Front Microbiol. 2023;14:1329330.doi:10.3389/fmicb.2023.1329330 [Pubmed: 38348304]
- Li F, Wang Y, Zhang Y, Shi P, Cao L, Su L, Zhu Q, Wang L, Lu R, Tan W, Shen J. Etiology of Severe Pneumonia in Children in Alveolar Lavage Fluid Using a High-Throughput Gene Targeted Amplicon Sequencing Assay. Front Pediatr. 2021; 9:659164.doi:10.3389/fped.2021.659164 [Pubmed: 34249808]
- Gaston DC, Miller HB, Fissel JA, Jacobs E, Gough E, Wu J, Klein EY, Carroll KC, Simner PJ. Evaluation of Metagenomic and Targeted Next-Generation Sequencing Workflows for Detection of Respiratory Pathogens from Bronchoalveolar Lavage Fluid Specimens. J Clin Microbiol. 2022 Jul 20;60(7):e0052622. doi: 10.1128/jcm.00526-22. Epub 2022 Jun 13. PMID: 35695488; PMCID: PMC9297812.
- Chiara M, D'Erchia AM, Gissi C, Manzari C, Parisi A, Resta N, Zambelli F, Picardi E, Pavesi G, Horner DS, Pesole G. Next generation sequencing of SARS-CoV-2 genomes: challenges, applications and opportunities. Brief Bioinform. 2021 Mar 22;22(2):616-630. doi: 10.1093/bib/bbaa297. PMID: 33279989; PMCID: PMC7799330.
- Thorburn F, Bennett S, Modha S, Murdoch D, Gunson R, Murcia PR. The use of next generation sequencing in the diagnosis and typing of respiratory infections. J Clin Virol. 2015 Aug;69:96-100. doi: 10.1016/j.jcv.2015.06.082. Epub 2015 Jun 18. Erratum in: J Clin Virol. 2015 Sep;70:128. PMID: 26209388; PMCID: PMC4533236.