More Information
Submitted: October 26, 2022 | Approved: November 04, 2022 | Published: November 07, 2022
How to cite this article: Razdan A, Arora R, Agarwal G, Sharma V, Singh N, et al. COVID-19 pandemic to endemic. Int J Clin Virol. 2022; 6: 043-049.
DOI: 10.29328/journal.ijcv.1001049
Copyright License: © 2022 Razdan A, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Neutralizing antibodies; Pathogenesis; Comorbid conditions; Immune system; Therapeutic Measures
COVID-19 pandemic to endemic
Alpana Razdan1*, Rajat Arora2, Gauri Agarwal3, Vandana Sharma1, Narendra Singh1, Jagdish Kandpal1, Sunil Tripathi1, Vijay Singh1, Saurabh Vishwakarma1 and Basudev Pal1
1Genestrings Diagnostics Centre, Malviya Nagar, New Delhi, India
2Yashoda Hospital and Research Centre, Nehru Nagar Ghaziabad, Uttar Pradesh, India
3Seeds of Innocence Maternity and Infertility Clinic, New Delhi, India
*Address for Correspondence: Alpana Razdan, Vice President & Head Lab Services, Genestrings Centre for Medical Genetics, 110017, New Delhi, India, Email: [email protected]
The COVID-19 pandemic appeared in late 2019 and became a major health concern with rapid transmission and very high mortality rates across the globe. Although precautionary, preventive, protective and therapeutic measures have been adopted against COVID-19, still the disease has drastically affected people. In order to overcome the challenges of the pandemic, the understanding of the route of transmission, its fusion with receptors and invasion into the human body and hacking the immune system, the viral genome was sequenced. The viral genome keeps on mutating and altering its original form into its subtypes. Moreover, age and comorbid conditions had their impact on developing the disease differing from individual to individual due to interaction varying between the host genome and virus. Considering the pathogenesis of the virus, neutralizing antibodies reduced the viral impact and severity. This review is focused on highlighting the COVID-19 genome, host genetic factors, the pathogenesis of the disease and available therapeutic measures to overcome the pandemic.
In December 2019, an alarming condition spread in Wuhan (China) and soon after the viral infection outbreak occurred as a pandemic across the globe about pneumonia of unknown cause. Later it became clear that the pandemic was caused by new severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) COVID-19 disease after the declaration of WHO (World Health Organization) [1] COVID-19 has drastically affected the overall economic, social and healthcare aspects of the world.
Seven different types of coronaviruses have been identified. The serious pathological problems and deaths were caused by three of these which are, severe acute respiratory syndrome coronavirus (SARS-CoV), the Middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV-2, the human coronavirus (HuCoV) 229E, HKU1, HCoV-NL63 and HCoV‐OC43, have been linked with mild common cold-like illness [2]. The SARS-CoV-2 variants and their types along with detailed information are available on the website of the Centre for Disease Control and Prevention [3].
Genetic architecture of COVID virus
COVID-19 was first reported in 2019 in Wuhan city of China. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) belongs to the family Coronaviridae, subfamily Coronavirinae and genus Betacoronavirus (the largest RNA viruses) and has been sequenced completely in previous reports [4]. The whole genome sequencing of COVID-19 has accelerated the integration of sequencing variants for accurate diagnosis and for developing vaccines and neutralizing antibodies and is available on the website of the National library of medicine [5].
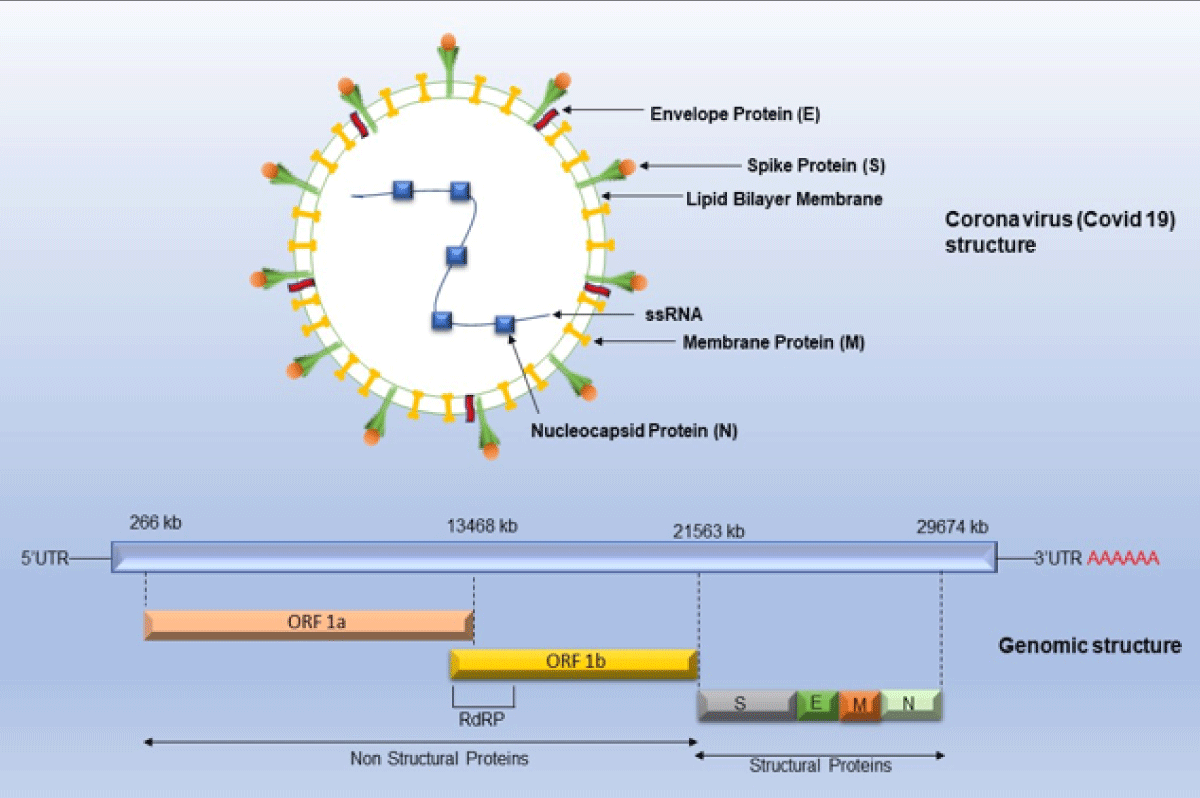
Multiple strategies were devised and implemented by scientists all over the world to standardize the sequencing protocol for SARS-CoV-2. The international workgroup known as Advancing Real-Time Infection Control Network (ARTIC), including scientists from the United Kingdom, Belgium, and the United States successfully designed a protocol of SARS-CoV-2 whole genome sequencing (WGS) on the Oxford Nanopore Technologies sequencing platforms and has been used widely in many labs [6]. The Office of Advanced Molecular Detection (AMD) at the Centre for Disease Control and Prevention (CDC) maintains a GitHub page with a comprehensive list of protocols and other tools used for SARS-CoV-2 whole genome sequencing on various platforms such as Illumina, PacBio and Ion Torrent [7,8]. The genome of the SARS-CoV-2 consists of approximately 30000 nucleotides and has been found to be more than 80% like previously reported human coronavirus (SARS-like Bat CoV) [9]. RNA virus COVID-19 has a high mutation rate because it does not have proofreading activity related to RNA-directed RNA polymerase and it is 2.5 × 10-6 substitutions per nucleotide per cell infection (s/n/c) [2]. A coronavirus contains six open reading frames (ORFs) that produce two polypeptides. The main structural proteins occur in 5/-3/ direction labeled as S (Spike), E (Envelope), M (Membrane) and N (Nucleoprotein) (Figure 1) [2].
Figure 1: Genomic structure representing single-stranded RNA, structural and non-structural proteins of SARS-CoV2./p>
The complete sequencing of the SARS-CoV-2 has helped in the development of RNA, DNA and peptide-based vaccines as well as attenuated viral vaccines to prevent the disease and disease-related severity [10]. Targeted therapeutic measures consist of antibodies to prevent virus entry, nucleotide analogs against viral replication and protease inhibitors to prevent virion formation and have been tested for their clinical efficacy previously [10].
Host genetic variants causing coronavirus patho-genicity
Numerous factors are linked to infection and disease pathogenicity including age, gender and comorbid conditions such as diabetes, hypertension, obesity and cardiovascular and cerebrovascular diseases. Males were found to be more prone to COVID-19 infection than females. The host genetic variants at the level of receptor site binding to the virus, immunogenic condition and inflammatory response via cytokines all may influence the disease risk and severity. The coronavirus pathogenicity varied from individual to individual depending upon ethnicity.
The variants in six genes of chromosome 3p21.31 including SLC6A20 (solute carrier transporter protein), LZTFL1, leucine zipper transcription factor-like 1), CCR9 (chemokine receptor type 9), FYCO1 (FYVE And Coiled-Coil Domain Autophagy Adaptor 1), CXCR6 (chemokine receptor 6), XCR1 gene (X-C Motif Chemokine Receptor 1) were reported to be associated with disease severity in a genome-wide association study (GWAS) in Spanish and Italian population [11]. The mortality-related gene variants include the ERAP2 (endoplasmic reticulum aminopeptidase 2) gene, the BRF2 (Transcription factor IIIB 50 kDa subunit) gene, the TMEM181 (Transmembrane Protein 181) gene and the ALOXE3 (Arachidonate Lipoxygenase 3) genes have three different alleles, which are the result of single nucleotide polymorphisms (SNPs) at restriction sites: rs150892504 and rs117665206, respectively. The immunogenetics mechanism involved in the pathogenesis of COVID-19 includes Human Leukocyte Antigens (i.e., HLA‐C*07:29 and HLA- B*15:27 alleles) mostly found to be associated with SARS‐CoV‐2 vulnerabilities in the Han population [12]. The four clinically relevant cases of severe coronavirus infection were screened via whole‐genome sequencing and the presence of loss‐of‐function mutations in the X‐chromosomal TLR7 (Toll Like Receptor) gene was reported, these were encoding for the TLR7, in COVID‐19 cases [13]. Several studies have documented multiple gene variants e.g., intercellular adhesion molecule 3, IFN‐γ (Interferon-gamma), MBL (mannose-binding lectin), IL4 (Interleukin-4), CCL2 (Chemokines), CCL5, Furin, TMPRSS2 (Transmembrane protease, serine 2) and CD209 (Cluster of Differentiation 209) promoter which may influence SARS‐CoV‐2 infection, varies markedly among different geographic populations, with the lowest incidence observed among Africans [14].
In a large study. Which was a part of the Million Veteran Program (MVP), SNP in MUC5B (Mucin 5B) genetic variant rs35705950‐T allele in the promoter region of chromosome 11 (11p15.5) was reported to be protective in function against COVID‐19 disease. The presence of the rs35705950‐T allele resulted in reduced cases of hospitalizations as well as lower episodes of pneumonia post‐COVID‐19 [15]. The reason is an increased expression of an rs35705950‐T allele in gene MUC5B in the lungs.
The angiotensin-converting enzyme insertion-deletion polymorphism (ACE1 ID) had been positively correlated with COVID-19 [16]. The TMPRSS2 gene variant rs383510 CC genotype played an important role as a predictor of a twofold increased risk of SARSCo-V2 [17]. This polymorphism affects the expression of encoded receptor protein and higher frequencies were found in the European and the American populations as compared to the Asian population.
Also, the variation in the ABO blood group, the ABO gene is located on chromosome 9 and encodes for enzyme glycosyltransferases responsible for the formation of antigens in blood type A and/or B. ABO blood group has an influence on the predisposition to COVID-19 infection, with the O and B blood groups having a reduced risk of infection [18]. Individuals harboring other than the O blood group have anti-A antibodies which makes them less susceptible to infection while other blood groups have been found at a higher risk of developing pulmonary embolism, thromboembolism in veins and thrombosis [19].
The ABO variation is related to vascular tone, leakage dysregulation, oxidative damage, cytokine storm production, and many other mechanisms. As a result, the ABO variant may/may not lead to COVID‐19 disease. The genetic variants/polymorphisms of the immune system have been detailed in a review by Adli, et al. [20].
In the first update on COVID-19 mapping of the genomic sequence was performed consisting of a genome-wide meta-analysis of 125,584 cases and more than 2.5 million individuals across 60 studies from 25 countries. This study found 23 genome-wide significant loci including a missense variant rs721917; A>G in the SFTPD gene of chromosome 10(10q22.3) associated with coronavirus disease phenotype and hospitalization [21]. The SFTPD is a pulmonary surfactant protein D (SP-D) gene that is involved in innate immune response, protecting the lungs against inhaled pathogens.
In a study focusing on comorbidities such as hypertension, diabetes and coronary artery disease, eleven common genes were reported, including TLR4, NLRP3 (NOD-like receptor gene), MBL2, IL6, IL1RN, IL1B, CX3CR1 (C-X3-C Motif Chemokine Receptor 1 gene), CCR5, AGT (Angiotensinogen), ACE and F2 (coagulation factor II) linked with COVID severity and infection [22]. These are mainly related to infection, inflammation, cytokine activity and receptor-binding process [22]. These genes along with their pathways have predictive value for determining the risk of infection and prognosis. These can also guide drug and novel targeted vaccine development [22].
Clinical manifestation of infection
The coronavirus clinical manifestation varies from individual to individual and in different age groups. The comorbid conditions example diabetes, heart disease, and the presence of any other disease. Children are less affected and the severity of the disease has not been observed as compared to adults and old people. The ACE-2 receptor expression decreases with age and might be a protective factor for reduced infection of the lungs in children. The ACE-2 being a member of the renin angiotensin-converting enzyme system has the functional properties of limiting pulmonary capillary leak and inflammation [23]. Children have strong innate immunity due to trained response (secondary to live vaccines) which prevented the infection at its site of entry at the time of exposure. Whereas adults were infected with higher and persistent viral loads and higher severity of disease manifestation. Adults have suppressed adaptive immunity not function in severe infection. Additionally, the children have excellent alveolar epithelium regeneration capacity effective against COVID-19 infection [23].
Mechanism of infection of viral hacking of the human immune system
COVID-19 or coronavirus has single-stranded RNA as a nucleic material, having the largest genome ranging from 26.4 kb - 31.7 kb in length [2]. The Structural proteins are encoded by the four structural genes, including spike (S), envelope (E), membrane (M) and nucleocapsid (N) genes (Figure 1). Finally, the host cells become virus factories and new virions are exported out from infected cells [24]. The initial step of the mechanism of SARS-CoV involves binding to coronavirus spike (S) proteins with aminopeptidase N (APN; HCoV-229E), angiotensin-converting enzyme 2 (ACE2; HCoV-NL63, SARS-CoV and SARS-CoV-2) and dipeptidyl peptidase 4 (DPP4; MERS-CoV) [25]. The S unit has a strong affinity for ACE receptors which are widely present in the respiratory system in the lungs, in the intestine, liver, heart, vascular endothelium, testis and in the ciliated epithelium of the kidney and in brain cells [25]. The S proteins are homotrimeric fusion glycoproteins divided into two subunits S1 and S2.S unit undergoes proteolytic cleavage by trypsin and furin at two sites between and S2. S1 receptor-binding domain (RBD) determines the pathogenicity and S2 has heptad repeat regions which are responsible for the fusion of viral and cellular membranes via the endosomal pathway and virus RNA is released into host machinery [26]. After that replication and transcription occur by replication and transcription complex.
The virus spreads via fomites and droplets and coughing, sneezing, and close contact between the infected and uninfected individual depending upon viral load. The symptoms include respiratory infections (flu-like to acute respiratory distress syndrome, pneumonia, cough and breathlessness), changes in taste receptors, shortness of breath, fever, dry cough, headache, pneumonia and even dyspnea. Some cases involve secondary infections, sepsis, organ failure, and gastrointestinal manifestations [27].
COVID variants having fast transmissibility such as delta (B.1.617.2), lambda (C.37), gamma (P1), beta (B.1.351) and alpha (B.1.1.7), were spread globally [28,29]. The major among these was infection spread by the delta variant and severity was high as compared to other subvariants.
The various variants of coronavirus are the Delta variant (21A Delta or B.1.617.2) and The Omicron variant (B.1.1.529). The various mutations have been reported due to substitutions at the positions P681R, L452R and T478K the transmissibility, pathogenicity and immune escape of the Delta variant. The additional mutations have been found at the positions T19R, R158G, T478K, D950N and deletions at the positions E156, F157-, A28271-, ORF8:D119- and ORF8:F120 [30-32]. The Omicron was highly transmissible due to its multiplication in the pulmonary cells about 70 times higher than the Delta variant [33]. Mutations in the Omicron combination of two mutations in the Spike protein (Q498 and N501Y) have been reported to increase the binding affinity to ACE2 receptors and have been documented in in vitro [34]. The increased immune evasion has been associated with the acquisition of an N-linked glycan in RBD (receptor binding domain) residue N370, resulting in neutralization escape to many antibodies [35].
Long covid and its effects on different organs and treatments
The patients were challenged by the long-term symptoms and other health complications caused by long COVID. The pandemic has caused an unprecedented escalation in morbidity as well as mortality in the initiation phase with 70% of survivors with medical complications. It has been estimated that among 30% - 70% of survivors of COVID infection long COVID symptoms including anxiety, headache, cough, cognitive impairment, shortness of breathing, pain in muscles, arthralgia, disturbed sleep and depression were observed. The respiratory epithelium was most affected and chronic inflammation, and airway dysfunction was observed among patients.
The SARS-CoV-2 infection lasts from a few weeks to months but its persistent symptoms post-COVID keep on lingering, this condition is known as ‘long COVID’. The pathophysiological manifestation of COVID has caused drastic changes in affected patients affecting multiple organs even in adults having low risk and without any comorbid condition. In a study from the United Kingdom, including two states and two hundred and one patients the most common symptoms, regardless of hospitalization observed were fatigue (98%), muscle ache (88%), shortness of breath (87%) and headache (83%). The evidence of Covid-2 affecting mild organs among patients reported was the impairment of the heart (32% ), lungs (33%), kidneys (12%), liver (10%), pancreas (17%) and spleen (6%) and hospitalization [36,37]. The study suggested that young patients with low risk had almost 70% impairments in one or more organs, four months post initial symptoms of SARS-CoV-2 infection.
The treatment and management of long COVID need a close follow-up and monitoring of clinical symptoms as the patients represent heterogeneously. The treatment is based on a holistic approach including assessment of pre-existing comorbid conditions and symptoms present at the time of case presentation to the clinician. As per the guidelines of the National Institute for Health and Care Excellence (NICE), the clinical investigation should be done as early as 4 weeks after initial acute symptoms [38]. Also, the National Institute of Health and Care Research (NIHR) also recommended that clinical management prioritize care for certain populations [39]. Therefore, a symptom-based approach was adopted for the management of long COVID and vaccination played an active role in significantly reducing the post COVID symptoms. Specific laboratory and clinical assessment tests were adopted to evaluate the clinical and physical impairment of long COVID -affected patients. To sum up, oxygen supplementation was provided for patients with dyspnoea, patients with cardiopulmonary symptoms underwent chest imagination and other investigations such as electrocardiography and spirometry, etc. Corticosteroids showed promising effects in pneumonia. Patient support and follow-up were also important to crop the situation perfectly.
Diagnosis
Accurate diagnosis has a pivotal role in the pandemic situation at the time when there was no alternative drug for COVID-19. RT-PCR is mainly used as a primary technique for detecting SARS-CoV-2. Several in-house and commercial kits have been on the market to diagnose the infection. Serological diagnosis is another method of detecting various antibodies e.g., IgG, IgM, and IgA in the serum of an infected patient [40]. Other advanced techniques e.g., lateral flow assay (LFA)-based assays like SHERLOCK (Specific High Sensitivity Enzymatic Reporter UnLOCKing), CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-Cas12a (AIOD-CRISPR) and FNCAS9 (using Cas9 from Francisella novicida) editor-limited uniform detection assay (FELUDA), etc. have produced promising results in rapid detection of pathogens [40]. Chronic persistence and reoccurrence of the disease have been observed among patients.
Therapeutic measures
In COVID-19 pandemic has caused drastic economic, social, and health-related changes. Initially, there was no treatment available for the virus and a huge number of patients died in the early 2020s across the globe. There was an urgent need of the hour to save the people. WHO launched guidelines and precautionary measures to prevent the infection. Effective prophylactic and treatment strategies were prepared to reduce the developmental time of vaccines, medicines neutralizing antibodies and other antiviral drugs.
With the non-availability of any immediate and targeted treatment, convalescent plasma was reported as a potential therapy for COVID-19 [41]. The COVID-19 patients. Post-hospitalization were transfused with convalescent plasma and successfully found to reduce the mortality rate [41-43].
The development of SARS-CoV-2 vaccines was accelerated across the world and many mRNAs and inactivated vaccines obtained emergency use for vaccination globally in order to control the spread of COVID-19 [43-45]. Neutralizing antibodies using effective vaccination served as the main treatment for COVID-19 due to their viral antigen-neutralizing efficiency [46]. Several drugs, such as hydroxychloroquine, arbidol and remdesivir, were reported safer and more effective. Several Indian traditional Ayurvedic medicines were claimed to boost immunity against COVID-19.
The pandemic has affected drastically every corner of the globe. The respiratory, heart system (arrhythmia and cardiac failure), gastrointestinal renal and brain systems (depression, myalgia and lack of sleep) are most susceptible to chronic effects post-COVID and the reason for its reoccurrence is still not clear at a molecular level even after vaccination. Post-COVID acute effects might be due to the persistence of cytokines. Monitoring disease severity, especially among patients having comorbid conditions and related complications is a part of the point of care of diagnosis and treatment.
The development of a safe and effective vaccine needs to successfully pass the critical phases of pre-clinical and clinical trials along with pharmacovigilance to avoid severe adverse effects [47]. International organizations including the World Health Organization (WHO), Coalition for Epidemic Preparedness Innovations (CEPI), Gavi alliance, Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV) and Bill and Melinda Gates Foundation (BMGF) amongst others had played an essential role to ensure adequate funding for vaccines with their collaborative approach against the spread of COVID-19 disease and to save the life [48]. In the 2021 year, there were 63 candidate vaccines in human clinical trials and more than 172 candidates in preclinical development across the Globe [49].
The vaccine development was accompanied by some challenges such as the type of vaccine antigen, immediate demand exceeding production, cold chain storage, etc. The antigen might be either an attenuated or inactive form of the whole virus or a part of it (either a protein or sugar). It can be an mRNA or DNA stimulating the host cells to produce neutralizing antibodies. Inactivated viruses, live attenuated viruses, and viral vectors can directly act on antigen-presenting cells (APC) to release antibodies against viral infection [50].
The state-of-the-art advanced technology including next-generation sequencing, mRNA delivery, expression techniques and novel recombinant vector systems had contributed to fastening the ‘next gen’ vaccine development and research.
Pangolin is a database (Phylogenetic Assignment of Named Global Outbreak LINeages), of dynamic nomenclature of SARS-CoV-2 lineages, known as the Pango nomenclature. With the help of this database, users can assign the SARS-CoV-2 genome sequence to the most likely lineage of SARS-CoV-2 query sequences [51]. For labs to use useful information from genome sequencing information of SARS-CoV-2 from the Pangolin database.
Considering the pandemic accelerating at an alarming rate with an unexpected number of deaths due to coronavirus, at the end of 2020, the Medicines and Healthcare Products Regulatory Agency (MHRA) in the UK, gave emergency approval to the world’s first COVID-19 vaccine, Pfizer/BioNTech’s BNT162b2 vaccine with active component tozinameran using mRNA delivery technology [52].
Soon after, the Moderna mRNA vaccine, Johnson & Johnson recombinant adenovector vaccine, and the Oxford-AstraZeneca adenovector vaccine, were sold under the brand names Covishield and Vaxzevria respectively (WHO), “World Health Organization” CDC, Centers for disease control and prevention. FDA, United States food and drug administration [52-56]. In Feb 2022, it was estimated that 61,2% of the world population was vaccinated with at least one dose of a vaccine, and in total, 10.14 billion doses had been administrated globally. Approximately 20,72 million doses/day were delivered [57]. Ten vaccines approved have been enlisted on the website COVID-19 Vaccine Tracker, 10 Vaccines Approved for Use by WHO, and have been reviewed in detail by Alexandridi, et al. [58].
Our laboratory Genestrings Diagnostic Centre Private Limited is one of the sentinel sites for genomic surveillance of COVID-19 detection and screening at IGI Airport New Delhi, India. Apart from locally recognized centers, we have played an active role during the pandemic outbreak, especially in testing coronavirus among international patients.
Pandemic shifting to endemic
The shifting of a pandemic to an endemic has a blurred borderline as the condition may convert at any time since the mutation in the virus and its emerging variants can reoccur. Although the vaccination drive has controlled the situation and transmission of the virus has been reduced significantly after adopting preventive and effective measures globally. Booster dose administration of the vaccine has been shown to restore the efficiency of neutralizing antibodies and hence prevention of COVID. The cases have reduced significantly and the normalized functional life predicts the endemic of COVID.
The shift from pandemic to endemic requires practical considerations including the breaking of the transmission chain of the virus, adopting stringent public health approaches, use of vaccination and on-time availability of therapeutics and managing the disease with proper health care infrastructure.
Global prevention and control measures for pandemic
The World Health Organisation (WHO) has recommended key guidelines for infection prevention and control (IPC). The IPC applied specific measures such as isolating the infected cohort, precautionary measures, providing specific training to curb the situation, applying universal masks, COVID-19 vaccination and implementing strategies for source control to stop the disease [59]. Several International and National organizations and healthcare workers have played a significant role to control the situation worldwide.
The pandemic is still crawling toward the endemic since we cannot predict the future situation. However, novel armamentariums using advanced techniques as well as pharmacological agents with effective neutralizing antibodies against SARS-CoV-2 have been developed and are in use to overcome the pandemic. Several therapeutics are in pipeline undergoing clinical trials which will ensure support to clinicians and public health and will be helpful in controlling any future pandemic with complete preparedness.
- World Health Organization. Emergencies. https://www.who.int/europe/emergencies/situations/covid-19.
- The SARS-CoV-2 genome: variation, implication and application. 2020. https://royalsociety.org/-/media/policy/projects/set-c/set-c-genome-analysis.pdf.
- Centers for Disease Control and Prevention. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html.
- Sah R, Rodriguez-Morales AJ, Jha R, Chu DKW, Gu H, Peiris M, Bastola A, Lal BK, Ojha HC, Rabaan AA, Zambrano LI, Costello A, Morita K, Pandey BD, Poon LLM. Complete Genome Sequence of a 2019 Novel Coronavirus (SARS-CoV-2) Strain Isolated in Nepal. Microbiol Resour Announc. 2020 Mar 12;9(11):e00169-20. doi: 10.1128/MRA.00169-20. PMID: 32165386; PMCID: PMC7067954.
- Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome. https://www.ncbi.nlm.nih.gov/nuccore/NC_045512.
- SARS-CoV-2 Sequencing Data: The Devil Is in the Genomic Detail. 2020. https://asm.org/Articles/2020/October/SARS-CoV-2-Sequencing-Data-The-Devil-Is-in-the-Gen.
- Hemarajata P. SARS-CoV-2 Sequencing Data: The Devil Is in the Genomic Detail. American Society for Microbiology. https://asm.org/Articles/2020/October/SARS-CoV-2-Sequencing-Data-The-Devil-Is-in-the-Gen.
- SARS-CoV-2_Sequencing. https://github.com/CDCgov/SARS-CoV-2_Sequencing.
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020 Mar;579(7798):265-269. doi: 10.1038/s41586-020-2008-3. Epub 2020 Feb 3. Erratum in: Nature. 2020 Apr;580(7803):E7. PMID: 32015508; PMCID: PMC7094943.
- Marian AJ. Current state of vaccine development and targeted therapies for COVID-19: impact of basic science discoveries. Cardiovasc Pathol. 2021 Jan-Feb;50:107278. doi: 10.1016/j.carpath.2020.107278. Epub 2020 Sep 2. PMID: 32889088; PMCID: PMC7462898.
- Severe Covid-19 GWAS Group, Ellinghaus D, Degenhardt F, Bujanda L, Buti M, et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N Engl J Med. 2020 Oct 15;383(16):1522-1534. doi: 10.1056/NEJMoa2020283. Epub 2020 Jun 17. PMID: 32558485; PMCID: PMC7315890.
- Lin M, Tseng HK, Trejaut JA, Lee HL, Loo JH, Chu CC, Chen PJ, Su YW, Lim KH, Tsai ZU, Lin RY, Lin RS, Huang CH. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med Genet. 2003 Sep 12;4:9. doi: 10.1186/1471-2350-4-9. PMID: 12969506; PMCID: PMC212558.
- Lu C, Gam R, Pandurangan AP, Gough J. Genetic risk factors for death with SARS‐CoV‐2 from the UK Biobank. medRxiv. 2020.
- Smatti MK, Al-Sarraj YA, Albagha O, Yassine HM. Host Genetic Variants Potentially Associated With SARS-CoV-2: A Multi-Population Analysis. Front Genet. 2020 Oct 2;11:578523. doi: 10.3389/fgene.2020.578523. PMID: 33133166; PMCID: PMC7567011.
- Verma A, Minnier J, Wan ES, Huffman JE, Gao L, et al. Million Veteran Program COVID-19 Science Initiative. A MUC5B Gene Polymorphism, rs35705950-T Confers Protective Effects Against COVID-19 Hospitalization but not Severe Disease or Mortality. Am J Respir Crit Care Med. 2022 Jun 30. doi: 10.1164/rccm.202109-2166OC. Epub ahead of print. PMID: 35771531.
- Verma S, Abbas M, Verma S, Khan FH, Raza ST, Siddiqi Z, Ahmad I, Mahdi F. Impact of I/D polymorphism of angiotensin-converting enzyme 1 (ACE1) gene on the severity of COVID-19 patients. Infect Genet Evol. 2021 Jul;91:104801. doi: 10.1016/j.meegid.2021.104801. Epub 2021 Mar 4. PMID: 33676010; PMCID: PMC7929788.
- Schönfelder K, Breuckmann K, Elsner C, Dittmer U, Fistera D, Herbstreit F, Risse J, Schmidt K, Sutharsan S, Taube C, Jöckel KH, Siffert W, Kribben A, Möhlendick B. Transmembrane serine protease 2 Polymorphisms and Susceptibility to Severe Acute Respiratory Syndrome Coronavirus Type 2 Infection: A German Case-Control Study. Front Genet. 2021 Apr 21;12:667231. doi: 10.3389/fgene.2021.667231. PMID: 33968142; PMCID: PMC8097083.
- Cheng Y, Cheng G, Chui CH, Lau FY, Chan PK, Ng MH, Sung JJ, Wong RS. ABO blood group and susceptibility to severe acute respiratory syndrome. JAMA. 2005 Mar 23;293(12):1450-1. doi: 10.1001/jama.293.12.1450-c. Erratum in: JAMA. 2005 Aug 17;294(7):794. Cheng, Yufeng [corrected to Cheng, Yunfeng]. PMID: 15784866.
- Trégouët DA, Heath S, Saut N, Biron-Andreani C, Schved JF, Pernod G, Galan P, Drouet L, Zelenika D, Juhan-Vague I, Alessi MC, Tiret L, Lathrop M, Emmerich J, Morange PE. Common susceptibility alleles are unlikely to contribute as strongly as the FV and ABO loci to VTE risk: results from a GWAS approach. Blood. 2009 May 21;113(21):5298-303. doi: 10.1182/blood-2008-11-190389. Epub 2009 Mar 10. PMID: 19278955.
- Adli A, Rahimi M, Khodaie R, Hashemzaei N, Hosseini SM. Role of genetic variants and host polymorphisms on COVID-19: From viral entrance mechanisms to immunological reactions. J Med Virol. 2022 May;94(5):1846-1865. doi: 10.1002/jmv.27615. Epub 2022 Feb 8. PMID: 35076118; PMCID: PMC9015257.
- COVID-19 Host Genetics Initiative. A first update on mapping the human genetic architecture of COVID-19. Nature. 2022 Aug;608(7921):E1-E10. doi: 10.1038/s41586-022-04826-7. Epub 2022 Aug 3. PMID: 35922517; PMCID: PMC9352569.
- Feng S, Song F, Guo W, Tan J, Zhang X, Qiao F, Guo J, Zhang L, Jia X. Potential Genes Associated with COVID-19 and Comorbidity. Int J Med Sci. 2022 Jan 24;19(2):402-415. doi: 10.7150/ijms.67815. PMID: 35165525; PMCID: PMC8795808.
- Dhochak N, Singhal T, Kabra SK, Lodha R. Pathophysiology of COVID-19: Why Children Fare Better than Adults? Indian J Pediatr. 2020 Jul;87(7):537-546. doi: 10.1007/s12098-020-03322-y. Epub 2020 May 14. PMID: 32410003; PMCID: PMC7221011.
- Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020 Mar 16;24:91-98. doi: 10.1016/j.jare.2020.03.005. PMID: 32257431; PMCID: PMC7113610.
- Cheng VC, Lau SK, Woo PC, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007 Oct;20(4):660-94. doi: 10.1128/CMR.00023-07. PMID: 17934078; PMCID: PMC2176051.
- Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu Rev Virol. 2016 Sep 29;3(1):237-261. doi: 10.1146/annurev-virology-110615-042301. Epub 2016 Aug 25. PMID: 27578435; PMCID: PMC5457962.
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 Feb 15;395(10223):497-506. doi: 10.1016/S0140-6736(20)30183-5. Epub 2020 Jan 24. Erratum in: Lancet. 2020 Jan 30;: PMID: 31986264; PMCID: PMC7159299.
- Campbell F, Archer B, Laurenson-Schafer H, Jinnai Y, Konings F, Batra N, Pavlin B, Vandemaele K, Van Kerkhove MD, Jombart T, Morgan O, le Polain de Waroux O. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill. 2021 Jun;26(24):2100509. doi: 10.2807/1560-7917.ES.2021.26.24.2100509. PMID: 34142653; PMCID: PMC8212592.
- Wink PL, Volpato FCZ, Monteiro FL, Willig JB, Zavascki AP, Barth AL, Martins AF. First identification of SARS-CoV-2 lambda (C.37) variant in Southern Brazil. Infect Control Hosp Epidemiol. 2021 Sep 2:1-2. doi: 10.1017/ice.2021.390. Epub ahead of print. PMID: 34470685; PMCID: PMC8564022.
- Kuzmina A, Wattad S, Khalaila Y, Ottolenghi A, Rosental B, Engel S, Rosenberg E, Taube R. SARS CoV-2 Delta variant exhibits enhanced infectivity and a minor decrease in neutralization sensitivity to convalescent or post-vaccination sera. iScience. 2021 Dec 17;24(12):103467. doi: 10.1016/j.isci.2021.103467. Epub 2021 Nov 15. PMID: 34805783; PMCID: PMC8591850.
- Saito A, Irie T, Suzuki R, Maemura T, Nasser H, Uriu K, Kosugi Y, Shirakawa K, Sadamasu K, Kimura I, Ito J, Wu J, Iwatsuki-Horimoto K, Ito M, Yamayoshi S, Loeber S, Tsuda M, Wang L, Ozono S, Butlertanaka EP, Tanaka YL, Shimizu R, Shimizu K, Yoshimatsu K, Kawabata R, Sakaguchi T, Tokunaga K, Yoshida I, Asakura H, Nagashima M, Kazuma Y, Nomura R, Horisawa Y, Yoshimura K, Takaori-Kondo A, Imai M; Genotype to Phenotype Japan (G2P-Japan) Consortium, Tanaka S, Nakagawa S, Ikeda T, Fukuhara T, Kawaoka Y, Sato K. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature. 2022 Feb;602(7896):300-306. doi: 10.1038/s41586-021-04266-9. Epub 2021 Nov 25. PMID: 34823256; PMCID: PMC8828475.
- Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, Planchais C, Porrot F, Robillard N, Puech J, Prot M, Gallais F, Gantner P, Velay A, Le Guen J, Kassis-Chikhani N, Edriss D, Belec L, Seve A, Courtellemont L, Péré H, Hocqueloux L, Fafi-Kremer S, Prazuck T, Mouquet H, Bruel T, Simon-Lorière E, Rey FA, Schwartz O. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021 Aug;596(7871):276-280. doi: 10.1038/s41586-021-03777-9. Epub 2021 Jul 8. PMID: 34237773.
- Lab studies, animal studies, and epidemiological data all indicate that Omicron may cause less severe disease than previous variants. Harvard Medical School. 2022. 〈https://www.health.harvard.edu/diseases-and-conditions/coronavirus-resource-cente.
- Zahradník J, Marciano S, Shemesh M, Zoler E, Harari D, Chiaravalli J, Meyer B, Rudich Y, Li C, Marton I, Dym O, Elad N, Lewis MG, Andersen H, Gagne M, Seder RA, Douek DC, Schreiber G. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat Microbiol. 2021 Sep;6(9):1188-1198. doi: 10.1038/s41564-021-00954-4. Epub 2021 Aug 16. PMID: 34400835.
- Nabel KG, Clark SA, Shankar S, Pan J, Clark LE, Yang P, Coscia A, McKay LGA, Varnum HH, Brusic V, Tolan NV, Zhou G, Desjardins M, Turbett SE, Kanjilal S, Sherman AC, Dighe A, LaRocque RC, Ryan ET, Tylek C, Cohen-Solal JF, Darcy AT, Tavella D, Clabbers A, Fan Y, Griffiths A, Correia IR, Seagal J, Baden LR, Charles RC, Abraham J. Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain. Science. 2022 Jan 21;375(6578):eabl6251. doi: 10.1126/science.abl6251. Epub 2022 Jan 21. PMID: 34855508; PMCID: PMC9127715.
- Lacobucci G. Long covid: Damage to multiple organs presents in young, low-risk patients. BMJ 2020; 371:m4470 | doi: 10.1136/bmj.m4470 1.
- Dennis A, Wamil M, Kapur S, Alberts J, Badley AD. Multi-organ impairment in low-risk individuals with long COVID. https://www.medrxiv.org/content/10.1101/2020.10.14.20212555v1.full.pdf.
- COVID-19 rapid guideline: managing the long-term effects of COVID-19. London: National Institute for Health and Care Excellence (NICE); 2020 Dec 18. PMID: 33555768.
- NIHR. LIVING WITH COVID19. London National Institute for Health Research. 2021.
- Islam KU, Iqbal J. An Update on Molecular Diagnostics for COVID-19. Front Cell Infect Microbiol. 2020 Nov 10;10:560616. doi: 10.3389/fcimb.2020.560616. PMID: 33244462; PMCID: PMC7683783.
- Chen L, Xiong J, Bao L, Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis. 2020 Apr;20(4):398-400. doi: 10.1016/S1473-3099(20)30141-9. Epub 2020 Feb 27. PMID: 32113510; PMCID: PMC7128218.
- Klassen SA, Senefeld JW, Johnson PW, Carter RE, Wiggins CC, Shoham S, Grossman BJ, Henderson JP, Musser J, Salazar E, Hartman WR, Bouvier NM, Liu STH, Pirofski LA, Baker SE, van Helmond N, Wright RS, Fairweather D, Bruno KA, Wang Z, Paneth NS, Casadevall A, Joyner MJ. The Effect of Convalescent Plasma Therapy on Mortality Among Patients With COVID-19: Systematic Review and Meta-analysis. Mayo Clin Proc. 2021 May;96(5):1262-1275. doi: 10.1016/j.mayocp.2021.02.008. Epub 2021 Feb 17. PMID: 33958057; PMCID: PMC7888247.
- Dong Y, Dai T, Wei Y, Zhang L, Zheng M, Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct Target Ther. 2020 Oct 13;5(1):237. doi: 10.1038/s41392-020-00352-y. PMID: 33051445; PMCID: PMC7551521.
- Shin MD, Shukla S, Chung YH, Beiss V, Chan SK, Ortega-Rivera OA, Wirth DM, Chen A, Sack M, Pokorski JK, Steinmetz NF. COVID-19 vaccine development and a potential nanomaterial path forward. Nat Nanotechnol. 2020 Aug;15(8):646-655. doi: 10.1038/s41565-020-0737-y. Epub 2020 Jul 15. PMID: 32669664.
- Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, Zaks T; COVE Study Group. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med. 2021 Feb 4;384(5):403-416. doi: 10.1056/NEJMoa2035389. Epub 2020 Dec 30. PMID: 33378609; PMCID: PMC7787219.
- Zhang Q, Xiang R, Huo S, Zhou Y, Jiang S, Wang Q, Yu F. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct Target Ther. 2021 Jun 11;6(1):233. doi: 10.1038/s41392-021-00653-w. PMID: 34117216; PMCID: PMC8193598.
- Sempowski GD, Saunders KO, Acharya P, Wiehe KJ, Haynes BF. Pandemic Preparedness: Developing Vaccines and Therapeutic Antibodies For COVID-19. Cell. 2020 Jun 25;181(7):1458-1463. doi: 10.1016/j.cell.2020.05.041. Epub 2020 May 27. PMID: 32492407; PMCID: PMC7250787.
- Sharma O, Sultan AA, Ding H, Triggle CR. A Review of the Progress and Challenges of Developing a Vaccine for COVID-19. Front Immunol. 2020 Oct 14;11:585354. doi: 10.3389/fimmu.2020.585354. PMID: 33163000; PMCID: PMC7591699.
- WHO. Draft landscape of COVID-19 candidate vaccines. https://www.who.int/ publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (World Health Organization, 2020).
- Zhang Z, Shen Q, Chang H. Vaccines for COVID-19: A Systematic Review of Immunogenicity, Current Development, and Future Prospects. Front Immunol. 2022 Apr 27;13:843928. doi: 10.3389/fimmu.2022.843928. PMID: 35572592; PMCID: PMC9092649.
- Phylogenetic Assignment of Named Global Outbreak Lineages. https://cov-lineages.org/resources/pangolin.html.
- COVID-19 advice for the public: Getting vaccinated. 2019. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/covid-19-vaccines/advice.
- World Health Organization. www.who.int
- Centers for Disease Control and Prevention. https://www.cdc.gov
- COVID-19 Bivalent Vaccine Boosters. https://www.fda.gov
- Livingston EH, Malani PN, Creech CB. The Johnson & Johnson Vaccine for COVID-19. JAMA. 2021 Apr 20;325(15):1575. doi: 10.1001/jama.2021.2927. PMID: 33646285.
- Coronavirus (COVID-19) Vaccinations. ourworldindata.org/covid-vaccinations.
- Alexandridi M, Mazej J, Palermo E, Hiscott J. The Coronavirus pandemic - 2022: Viruses, variants & vaccines. Cytokine Growth Factor Rev. 2022 Feb;63:1-9. doi: 10.1016/j.cytogfr.2022.02.002. Epub 2022 Feb 12. PMID: 35216872; PMCID: PMC8839804.
- WHO policy brief: Maintaining infection prevention and control measures for COVID-19 in health care facilities. 2022. https://www.who.int/publications/i/item/WHO-2019-nCoV-Policy_Brief-IPC-2022.1