More Information
Submitted: April 14, 2021 | Approved: April 27, 2022 | Published: April 28, 2022
How to cite this article: Oliveira AAS, Silva AMP, da Silva Queiroz JA, de Souza PRF, Salcedo JMV, et al. ACE2 and TMPRSS2 polymorphisms and the development of COVID-19: a review of the literature. Int J Clin Virol. 2022; 6: 017-023.
DOI: 10.29328/journal.ijcv.1001044
Copyright License: © 2022 da Silva Oliveira AA, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Polymorphisms; ACE2; TMPRSS2; COVID-19; Susceptibility
ACE2 and TMPRSS2 polymorphisms and the development of COVID-19: a review of the literature
Adrhyan Araújo da Silva Oliveira1,4, Ana Maisa Passos da Silva1,2, Jackson Alves da Silva Queiroz1,2, Paulo Ricardo Freitas de Souza1,4, Juan Miguel Villalobos Salcedo3 and Deusilene Souza Vieira1-4*
1Laboratório de Virologia Molecular, Fundação Oswaldo Cruz Rondônia, FIOCRUZ/RO, Porto Velho, RO, Brazil
2Programa de Pós-Graduação em Biologia Experimental, Universidade Federal de Rondônia, UNIR, Porto Velho, RO, Brazil
3Universidade Federal de Rondônia, UNIR, Porto Velho, RO, Brazil
4Centro de Pesquisa em Medicina Tropical, CEPEM, Porto Velho, RO, Brazil
*Address for Correspondence: Deusilene Vieira, Laboratório de Virologia Molecular, Fundação Oswaldo Cruz Rondônia, Fiocruz/RO, Porto Velho, RO, Brazil, Email: [email protected]
SARS-CoV-2 is a virus that has a positive-sense, single-stranded RNA genome that encodes 4 structural proteins, the main one being the S protein (Spike) responsible for mediating with ACE2 and TMPRSS2 for entry into the host cell. The study of single nucleotide polymorphisms (SNPs) of ACE2 and TMPRSS2 can elucidate their possible intervention in the action of the protein, its activity, and the gene expression of encoding these enzymes, which may increase susceptibility to viral infection. From this, literature searches were carried out until December 2021, listing 11,820 publications for literary analysis on the described genetic variations of these protein structures, as well as their relation and influence on the pathology. It was possible to conclude that there is a great influence exerted by genetic variability in ACE2 and TMPRSS2 increasing the ability of the virus to bind to the host cell and the development of COVID-19 with complications.
In late 2019, in Wuhan in China, 27 cases of pneumonia of unknown cause were reported, subsequently concluding that the pathology was caused by a new type of Coronavirus with a high capacity for human-to-human transmission [1]. In February 2021, the International Committee on Viral Taxonomy (ICTV) named this virus Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), and the World Health Organization (WHO) identifies the disease it causes as COVID-19, elevating the disease status to a pandemic in March 2020 based on its high transmission rate and rapid global spread [1,2].
COVID-19 is an acute respiratory disease characterized by flu-like symptoms, where Severe Acute Respiratory Syndrome (SARS) is the aggravation of this pathology [3]. Since the beginning of the pandemic, a higher prevalence and development of the disease is observed in the elderly and people with comorbidities, however, there is the visualization of different clinical pictures among individuals who present similarities in age and sex, where some develop the symptoms of the disease, with worsening regardless of comorbidities, as well as cases of infected individuals with asymptomatic picture [4].
With the beginning of vaccination, it was possible to establish control over the incidence of the pathology in the world, and as a consequence, there were drops in its prevalence and mortality rates [5,6]. Cases of SARS-CoV-2 infection and the development of the disease are recurrently visualized even in immunized individuals, with the recommendation of the interposition of booster doses to ensure greater durability of the acquired immune response [6] .
SARS-CoV-2 infection often results in the development of COVID-19 and for the viral infection process to be successful there must be the interaction of the viral particle with specific human cellular receptors such as Angiotensin-Converting Enzyme 2 (ACE2) [7]. And in some cases, other proteins are involved, such as Transmembrane Protease Serine 2 (TMPRSS2) [3,8].
Disease susceptibility factors such as environment, climate, comorbidities, and especially the genetics of individuals are intrinsically linked [9]. Each and every structure of the body is encoded by specific genes present in the genetic material, each gene having combinations of alleles that configure different structures. Point mutations are known as Single Nucleotide Polymorphisms (SNPs) play a genotypic role in the phenotypic diversity of diseases [10].
Thus, the study of the SNPs of ACE2 and TMPRSS2 may be crucial for understanding the greater susceptibility to viral infection in some individuals and from this to elucidate possible causes of disease development and presentation of different symptoms even in similar individuals and in vaccinated individuals [3]. Therefore, understanding the functional and genetic role of ACE2 and TMPRSS2 and how their polymorphisms may influence the greater susceptibility to viral infection and the development of the pathology is of great importance.
Data source and strategy
Searches were conducted on the PubMed, PubMed Central, Gene, GEO DataSets, and Genome platforms of the National Center for Biotechnology and Information (NCBI) to identify relevant articles on the subject. With this, keywords related to COVID-19 and SARS-CoV-2 infection correlated to ACE2 and TMPRSS2 were used with an emphasis on their polymorphisms and their links to viral virus infection in humans. Clinical progression, severity, risk factors, and vaccination are research topics also addressed for inclusion criteria in this review.
Study selection and eligibility requirements
Observational studies (cross-sectional, cohort, and case-control studies) were selected with the exception of case reports and case series. From this, research reporting the importance of ACE2 and TMPRSS2 in SARS-CoV-2 viral infection was included, as well as research addressing polymorphisms in genes encoding these proteins involved in virus-infected individuals, the development of the disease, and increased susceptibility to infection as a consequence of genetic variability in the host.
Study identification and selection
Searches were conducted until December 2021, from this, 11,820 citations were identified (PubMed: 10,580; PubMed Central: 956; Gene: 99; Geo DataSets: 30; Genome: 155), from these studies articles were excluded based on their titles, abstract, relevance to the study, duplicates and study type (N=11,557), from the 263 remaining 224 did not meet the eligibility criteria. With this, 39 were selected for full-text evaluation. All studies comprised observational research and all were matched with the proposed inclusion criteria.
Viral genomics of SARS-CoV-2 and its infectious process
SARS-CoV-2, belonging to the genus Betacoronavirus (group 2) and to the family Coronavidae [11,12], is an enveloped virus that has as genome a ≊ 30-kb positive-sense single-stranded RNA that encodes 4 structural proteins being N (nucleocapsid), M (Membrane), E (Envelope) and S (Spike) and 16 nonstructural proteins [2,4,12]. Being a novel beta coronavirus, SARS-CoV-2 has a genome with ≅ 80% similarity to the SARS-CoV genomic sequence and 50% to MERS-CoV [2,4,13], because of this, its structural proteins share more than 90% similarity with those of SARS-CoV [2,4] differing only from Protein S in total amino acid size, sequence similarity and receptor-binding domain (RBD) [2,4,5,13].
Like SARS-CoV, the viral infection process by SARS-CoV-2 depends on the interaction of the S protein in its “open” state with ACE2 [2,4,7,11]. After binding the RBD domain to ACE2, the Spike protein undergoes proteolytic activation mediated by host cell proteases such as FURIN or TMPRSS2, which cleave between the S1 and S2 subunits of the viral protein [3,12-15]. This process induces the fusion of the viral envelope with the cell membrane which results in the internalization of the genome into the cytosol [4,14,15]. Another method of entry of the viral particle into the cell is through receptor-mediated [14,15] where interaction with ACE2 induces an invagination of the cell membrane and the creation of an endosome[8]. Some proteases can act in this process such as Cathepsin B and L [8,14] inducing fusion of the viral envelope with the endosomal membrane and release of the viral genome into the cytoplasm [2,15].
In the cell cytoplasm, the Open Reading Phases (ORFs) 1a and ORF1ab of the viral genome will be translated by the host cell ribosomes into two polyproteins (pp1a and pp1ab) [4,15,16]. Translation products are cleaved by the viral proteases PLpro and Mpro into nonstructural proteins (Nsp) 1 to 16 [2,16], which will later assemble to form the Replication-Transcription Complex (RTC) in the host cell[2][4][16][17]. With the formation of the RTC, the ssRNA(+) strand will serve as a template for the synthesis of the negative-sense genomic RNA (ssRNA-) [2,17] and for the transcription of subgenomic mRNAs which will be translated into structural and accessory proteins. The ssRNA(-) in turn will be used for the synthesis of positive genomic RNA for the assembly of new viral particles [2,16,18]. The E, M, and S proteins are directed to the endoplasmic reticulum where they will be activated by proteolytic cleavage, except for the S protein [12,15-17], while the N protein targets the positive RNA strand and binds to form the nucleoprotein complex [1,2,16,17]. From there, the nucleocapsid and structural proteins move to the Endoplasmic Reticulum-Golgi Intermediate Compartment (ERGIC) for the assembly of new virions [2,16]. After this process and its maturation, vesicles are formed that carry the viral particle to the extracellular environment and release it by exocytosis [2,14,16,17,19].
Angiotensin-converting enzyme 2 (ACE2) a protein with vascular regulatory function
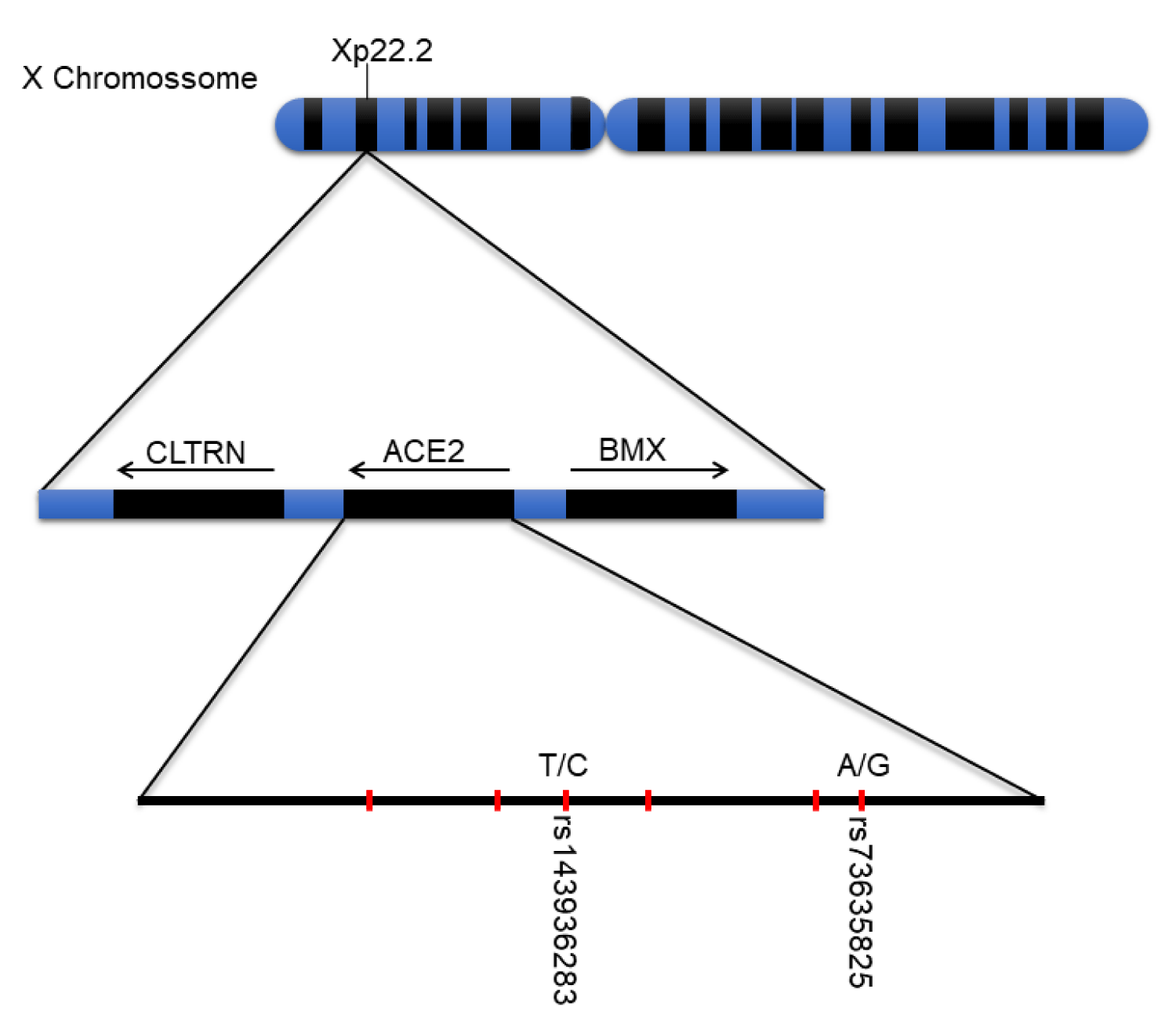
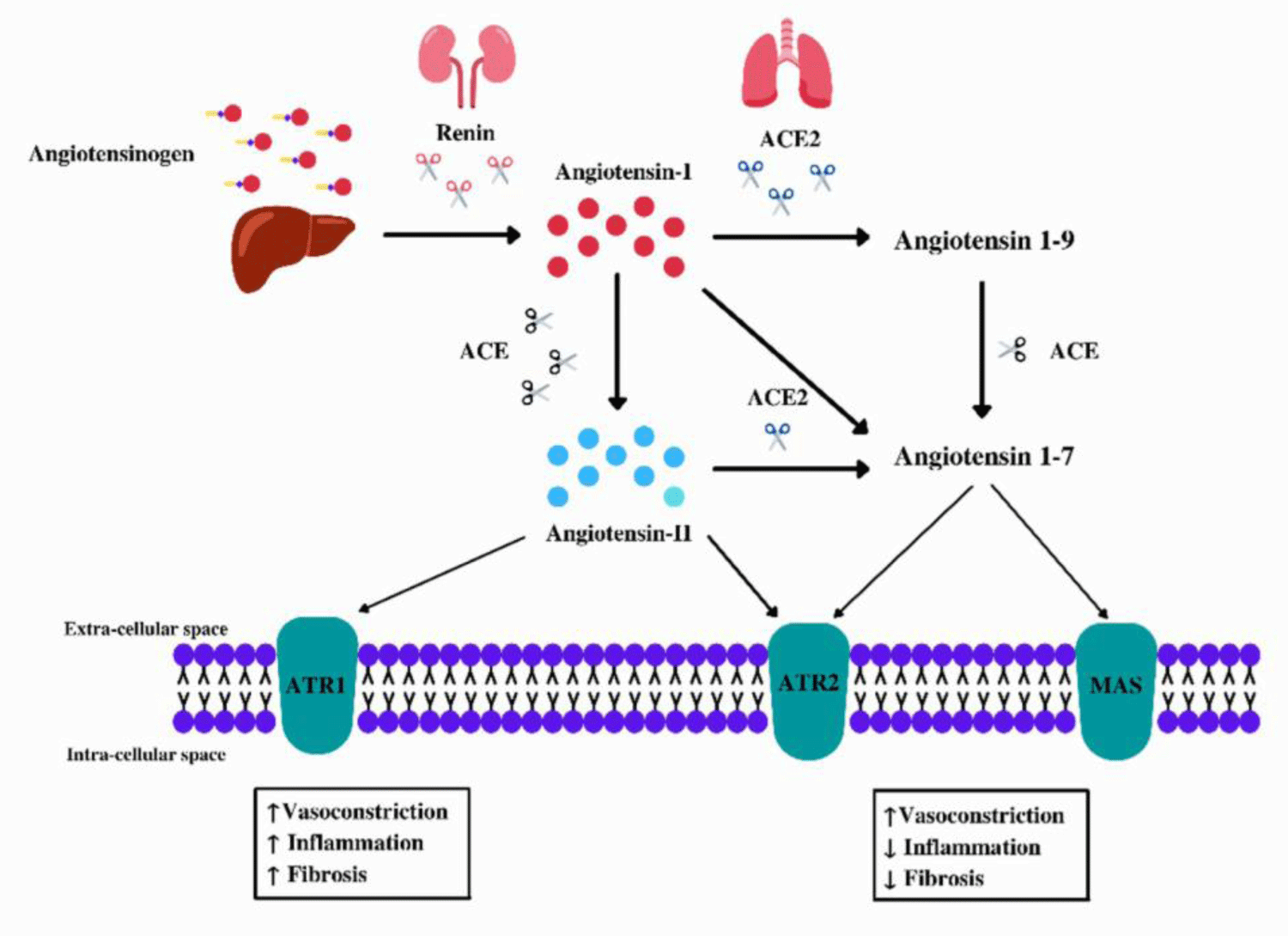
Angiotensin-converting enzyme 2 (ACE2) is a protein responsible for cleaving angiotensin I into angiotensin 1-9, and angiotensin II into the vasodilator angiotensin 1-7, which is encoded by the ACE2 gene, located on sex chromosome Xp22.2 (Figure 1) [7,11]. The protein is part of the set of enzymes participating in theRenin-Angiotensin-Alderone System (RAAS), which plays a major role in the regulation of the cardiovascular system, affecting blood pressure, and sodium regulation, fluid balance, and vascular function [11].
Figure 1: X chromosome, Xp22.2 gene, and important ACE2 polymorphisms. Subtitle: Main polymorphisms and their locations on the X chromosome and ACE2 gene.
The expression of this gene in the human body is mainly distributed in organs and cells that play roles in controlling cardiovascular, renal, and gastrointestinal function, but it can also be found in other tissues of the human body [11].
ACE2 and its function in SARS-CoV-2 viral infection
ACE2 also plays an associative role in the development of COVID-19, playing an essential role in the process of viral infection by the Severe Acute Respiratory Syndrome-causing Coronavirus (SARS-CoV-2), mediating the entry of the viral particle into the host cell through the interaction of the receptor-binding domain (RBD) of the surface glycoprotein Spike (S) (located on the viral envelope) with ACE2 [12]. The process triggered by the internalization of ACE2 leads to decreased physiological protein action, impairing the processes dependent on this enzyme [11].
The divergence of enzyme expression between individuals is a factor of irrefutable importance for understanding the risks of COVID-19, factors such as gender and age influence the higher or lower amount of ACE2 [20,21]. Gene expression assays with humans and mice have shown that there is a higher detection of the protein-coding mRNA in elderly individuals, especially males, where there is an excessive increase of the protein structure expression in the lungs and kidneys of these individuals [22]. In turn, females, despite having two X chromosomes each with a functional copy of the ACE2 gene, have not been detected to increase the protein in their organs due to a complex transcriptional and proteolytic control and hormonal factors that counteract the overexpression of the enzyme in women [20-23].
In theory, the low expression of this protein would be beneficial to combat SARS-CoV-2 by inhibiting its infection mechanism, but there is no data that proves the effectiveness of this measure without harm to the human organism [22]. After the viral particle binds to ACE2, there is the internalization of this protein, which leads to a decrease in its physiological action and damage to the RAAS (Figure 2) [11,12,22,24]. The absence of ACE2 in mice was associated with lung damage after viral infection [22], there is also a correlation between increased organ damage and the development of the severe condition of COVID19 [5], in addition to the positive regulation of Ang II in humans due to ACE2 deficiency [11]. Therefore, decreased expression of ACE2 may contribute to the control of SARS-CoV-2 viral infection, however, as a consequence it will cause delayed conversion of Ang II to angiotensin 1-7 and thus increased interaction with ATR1 resulting in tissue damage [11,24].
Figure 2: Renin-Angiotensin Aldosterone System (RAAS). Subtitle: Angiotensinogen is secreted by the liver, then the adrenal glands secrete the enzyme Renin which cleaves it into Angiotensin I (ANG I), subsequently Ang I can be cleaved either by ACE → Ang II or by ACE2 which can transform it into angiotensin 1-9 or angiotensin 1-7. SARS-CoV-2 can promote a higher concentration of Ang II, as the interaction with ACE2 internalizes it in the cytoplasm of the cell preventing it from performing its physiological function, with this Ang II can bind to ATR1 causing hypertension.
ACE2 polymorphisms involved in increased suscepti-bility to COVID-19
Genetic variation influences increased susceptibility to developing COVID-19 [10,13], single nucleotide poly-morphisms (SNPs) characterize allele changes that can configure differences in the regulation of the expression of genes encoding these structures and in their respective functions in individuals of a population [10]. The ACE2 gene has a high degree of genetic polymorphisms with more than 1700 variations identified, a portion is used for understanding the increased susceptibility to viral infection by SARS-CoV-2 in the molecular interaction with spike protein [3,12], among the associated SNPs, rs73635825 [3] and rs143936283 [25] have been shown to be variations of clinical importance.
The polymorphisms of ACE2 differ according to the allele frequency characterizing in some cases an influence on the RAAS, there is an association of SNPs of the protein with the development of several diseases such as hypertension, left ventricular hypertrophy, myocardial infarction [11,25]. With this, some described variations directly interfere with the surface interaction of ACE2 with angiotensinogen [25], it is important to note that dysfunction in this system can lead to cardiac and renal complications, since it covers vascular flow and sodium and hydric control [11], therefore, genetic variation is directly linked to increased susceptibility and to the development of the disorder COVID19 with complications.
Transmembrane Protease Serine 2 (TMPRSS2) a pro-tease of importance
The process of protein degradation is necessary for the life-sustaining cellular proteins [14], for that, they are submitted to constant renewal procedures (apoptosis) that require new protein synthesis and subsequent degradation, this process requires the action of proteins called proteases that have as function the hydrolysis of peptide bonds such as TMPRSS2 [11,14].
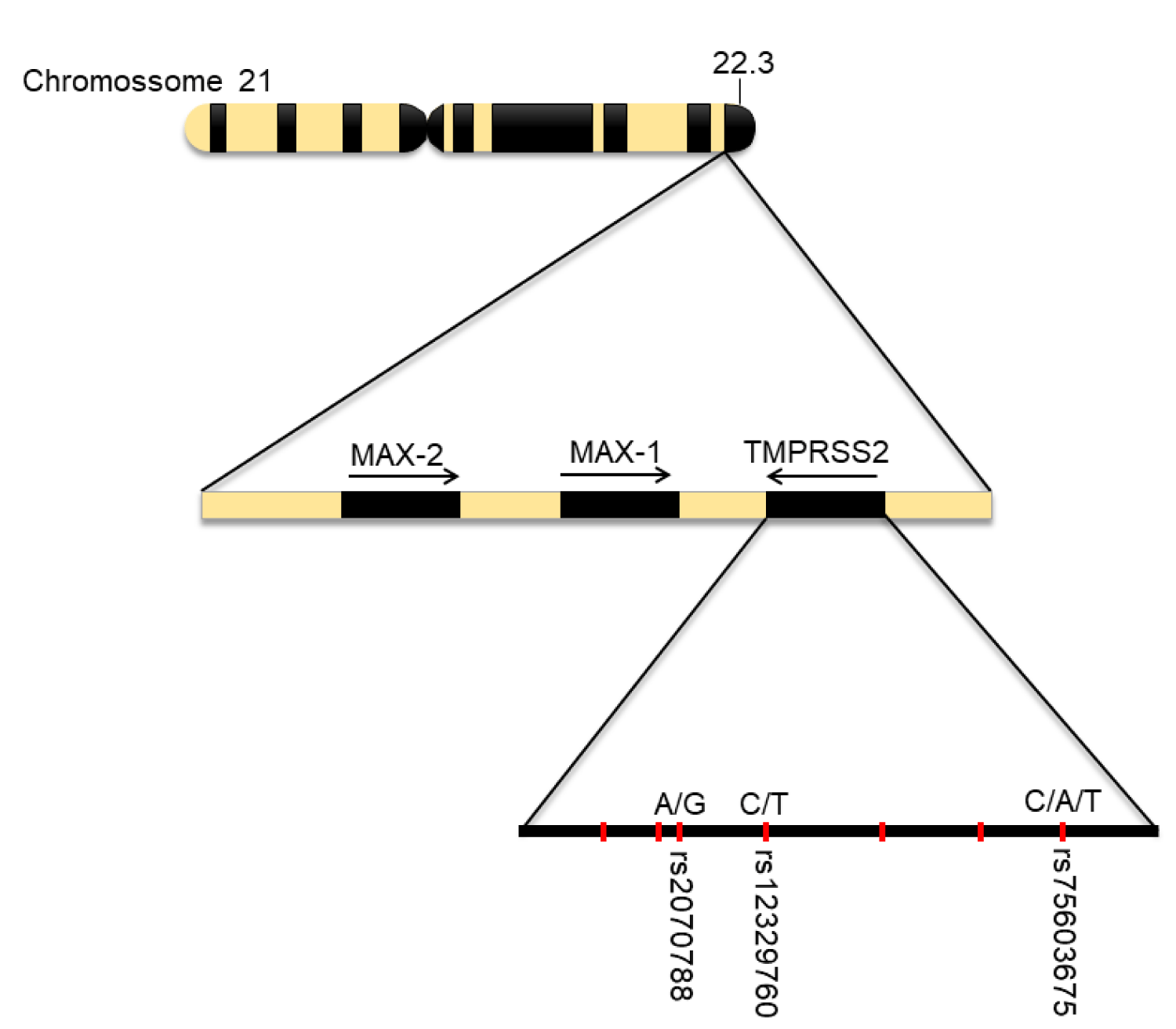
Transmembrane protease serine 2 (TMPRSS2) is a protein encoded by the TMPRSS2 gene, located on autosomal chromosome 21q22.3 [14]. Just like other serine proteases their physiological function is not yet fully elucidated, but there is their possible participation in indigestion, coagulation, embryonic development, and spermatogenesis [14]. Its expression was initially associated with the small intestine, however, it was later found to be more expressed in the prostate epithelium [14,26], because of this, there is an association of protease in carcinogenesis due to overexpression in the development of prostate cancer by molecular alterations and the formation of the TMPRSS2-ERG arrangement, where one TMPRSS2 allele loses its promoter and one of the ERG alleles receives it [27].
TMPRSS2 has a type II transmembrane domain, A domain, scavenger receptor, and serine protease domain [14], possibly its functionality would be related to a cell-specific ligand mediating signal receptor [14,19]. It can be found in the nasal epithelium, trachea, epithelial cells of the bronchi, digestive tract, and prostate, its expression is developmentally regulated and increases by the age range of individuals [12,14,27,28].
TMPRSS2 and its role in SARS-CoV-2 viral infection
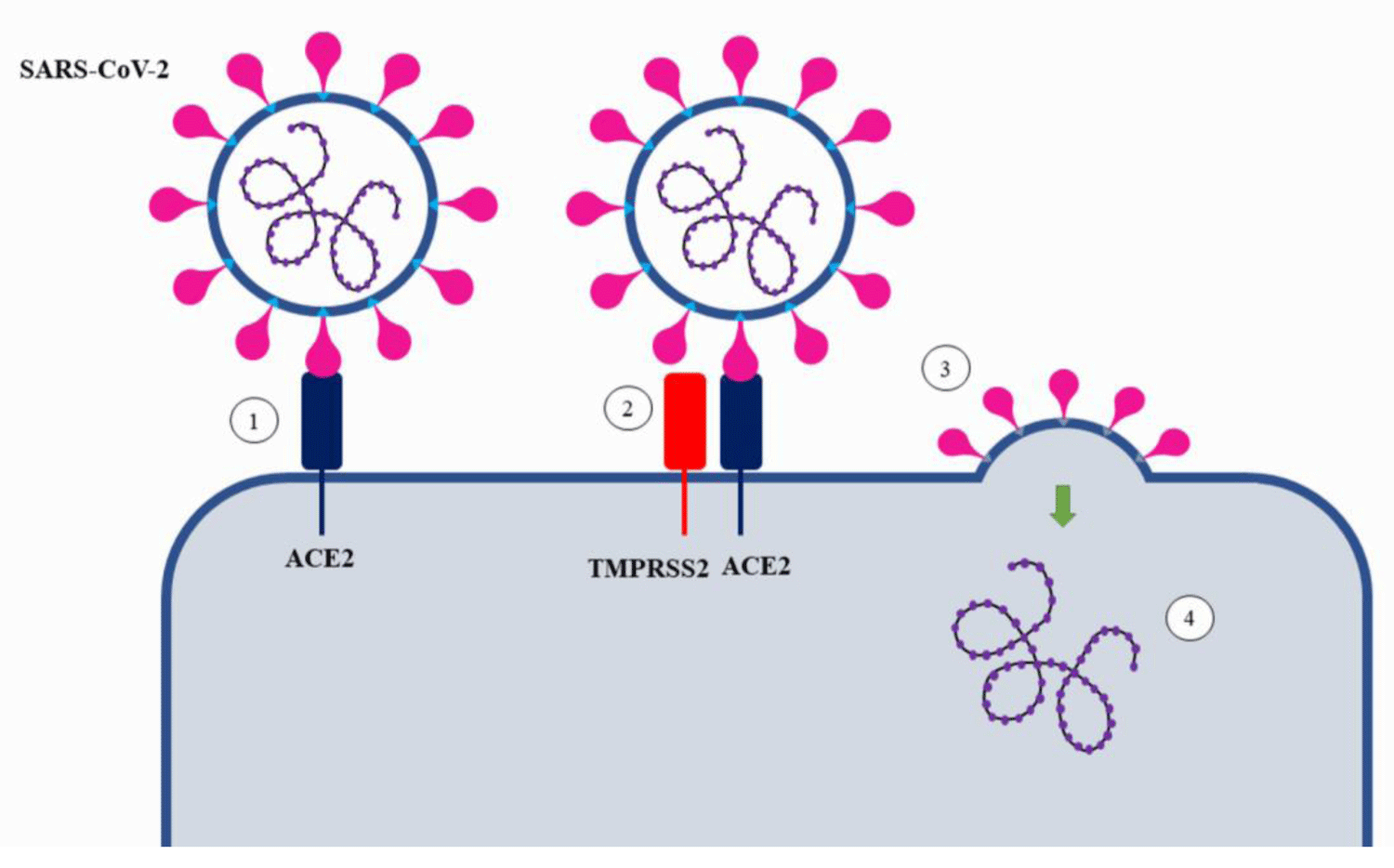
Of the 4 structural proteins encoded in the SARS-CoV-2 viral genome[19], the Spike protein is the main one for acting in the process of binding the viral particle to the host cell (Figure 3) [4,12,19]. The S protein consists of two subunits: S1 - acts as a binding region for ACE2; S2 - acts in promoting membrane fusion and proteolytic action [4,11]. At the end of the replicative cycle of the virus the new viral particles leave the host cell with the S protein not activated [4], therefore, the process of virus penetration is intrinsically dependent not only on the angiotensin-converting enzyme 2 but also on the participation of proteases such as TMPRSS2 [4,15,29].
Figure 3: Cleavage of the S protein of SARS-CoV-2 by TMPRSS2. Subtitle: 1) Viral particle approaches and interacts with angiotensin-converting enzyme 2 (ACE2); 2) TMPRSS2 cleaves the S2’ subunit of the Spike protein by activating its RBD domain and interacting with ACE2; 3) Fusion of the viral envelope induced by activation of the virus S protein and release of the viral genome into the cytoplasm.
Like other Coronaviruses, SARS-CoV-2 uses two steps for entry into target cells [4,11,12], first, the viral particle binds to ACE2 on the surface of the host cell membrane, then the S-protein is cleaved between the S1 and S2 subunits by endogenous proteases in the human body [15] as the transmembrane serine protease 2, it is important to emphasize that the cleavage must be performed after the binding of the virus to the surface protein, if it occurs earlier there is the possibility of inactivation of the Sprotein [14], thus, even though there is a large availability of proteases in the membrane, their mobility, and non-permanence in the vicinity of ACE2 besides the possibility of inactivation of the spike protein before the interaction process are factors that justify their disuse [15]. Hence, the location and time of action of proteases such as TMPRSS2 are important for viral infection and pathogenicity [14,15].
In addition to transmembrane serine protease 2, other proteases may be involved in cleavage and activation of protein S such as Cathepsin B and L in the case of receptor-mediated endocytosis [8,14], therefore, individuals who do not have large amounts of TMPRSS2 expression may use them as mediators of viral envelope protein activation and fusion with the endosome membrane[8]. Furin is another protease widely used for cleavage of the S protein of SARS-CoV-2 and is often associated with TMPRSS2 replacement in case of its absence [8,30].
TMPRSS2 polymorphisms associated with increased susceptibility to COVID-19
The genetic variability of TMPRSS2 is directly related to increased susceptibility to COVID-19 [9,31], the rs75603675 and rs12329760 (Val160Met) polymorphisms of TMPRSS2 (Figure 4) have great clinical importance, studies identify that the variation encoding Val160Met is identified in much of the world population with higher allele frequency ≊ 25% and especially for EAS (Overpopulation Code for East Asia) with an allele frequency of ≊ 40% [31-33]. These more prevalent SNPs on TMPRSS2 offer potential effects for greater differential genetic disposition on COVID-19, as well as, potentiating the development of risk factors such as prostate cancer in males, therefore, the study of oncogenic factors [9,26,32].
Figure 4: Human Chromosome 21, gene 21q22.3 and TMPRSS2 and COVID-19 associated polymorphisms. Subtitle: Main polymorphisms and their locations on chromosome 21 and TMPRSS2 gene.
The potential action of TMPRSS2 on COVID-19 may be expressed in the rs2070788 and rs383510T polymorphisms, which are associated with higher protease expression in human lung tissue [31]. Similar to the above, most studies highlight a higher risk for males linked not only to polymorphisms but also to androgen disposition, an observation based on hair loss related to COVID19 male subjects admitted [15,34,35]. Interestingly, the TMPRSS2 promoter gene has a 15bp AR (Androgen Receptor) response element and the human protease appears to be regulated by these receptors, therefore indicating gender differences in SARS-CoV-2 infection [32].
Moreover, the gene encoding TMPRSS2, being located on autosomal chromosome 21q22.3, may in thesis verify a higher risk of infection by SARS-CoV-2 in individuals with trisomy 21 (Down Syndrome) [9]. Increased expression of protease may be potentiation of the infection factor [9,32,36]. Studies have identified an increase in the presence of the enzyme in both mice and humans as they age[4], therefore, identifying that possible regulation of TMPRSS2 development may result in increased protection in infants and children [27,37,38], thus, the study of age-related polymorphisms of this protein may be of great interest [31,35,39].
The ACE2 and TMPRSS2 polymorphisms were shown to be great indicators of greater susceptibility to COVID-19, since their variations not only affect the viral ability to infect the cell due to overexpression in some cases, but also the development of other pathologies due to the internalization of these proteins, as in the case of ACE2, causing a decrease in its action and, in some cases, due to allelic variants, even the loss of affinity in the catalytic function, impairing extremely important physiological processes, such as RAAS. Furthermore, there is a need for other studies that address these genetic variations in isolated groups such as children, infants, and people with Down syndrome to study the possible control of expression of genes encoding these structures in a non-harmful way.
The in-depth study of the genetic aspects involved may elucidate a new pathway for new therapies for COVID-19; the importance of the knowledge of the genetic profile of individuals infected by the virus indicates a high potential for the discovery of new mechanisms involved in the greater vulnerability to viral infection, such as the polymorphisms cited. Moreover, a more cautious look is needed regarding the studies related to the male population, once it is demonstrated that factors such as age, age range, and genetic variation can directly affect individuals, not only for COVID-19 but also with the development of other complications such as infertility, prostate cancer, and cardiac complications. Thus, genetic knowledge proves to be a strong ally in understanding and combating SARS-CoV-2.
Authors’ contributions
Conceptualization: Deusilene Souza Vieira.
Data processing and analysis: Adrhyan Araújo da Silva Oliveira; Ana Maísa Silva Passos.
Methodology: Adrhyan Araújo da Silva Oliveira; Ana Maísa Silva Passos.
Writing–original draft: Adrhyan Araújo da Silva Oliveira; Ana Maísa Silva Passos; Jackson Alves da Silva Queiroz.
Writing–review and editing: Deusilene Souza Vieira; Juan Miguel Villalobos Salcedo; Ana Maísa Silva Passos; Jackson Alves da Silva Queiroz; Paulo Ricardo Freitas de Souza.
- Muralidar S, Ambi SV, Sekaran S, Krishnan UM. The emergence of COVID-19 as a global pandemic: Understanding the epidemiology, immune response and potential therapeutic targets of SARS-CoV-2. Biochimie. 2020 Dec;179:85-100. doi: 10.1016/j.biochi.2020.09.018. Epub 2020 Sep 22. PMID: 32971147; PMCID: PMC7505773.
- Arya R, Kumari S, Pandey B, Mistry H, Bihani SC, Das A, Prashar V, Gupta GD, Panicker L, Kumar M. Structural insights into SARS-CoV-2 proteins. J Mol Biol. 2021 Jan 22;433(2):166725. doi: 10.1016/j.jmb.2020.11.024. Epub 2020 Nov 24. PMID: 33245961; PMCID: PMC7685130.
- Singh H, Choudhari R, Nema V, Khan AA. ACE2 and TMPRSS2 polymorphisms in various diseases with special reference to its impact on COVID-19 disease. Microb Pathog. 2021 Jan;150:104621. doi: 10.1016/j.micpath.2020.104621. Epub 2020 Dec 2. PMID: 33278516; PMCID: PMC7709597.
- Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021 Mar;19(3):141-154. doi: 10.1038/s41579-020-00459-7. Epub 2020 Oct 6. Erratum in: Nat Rev Microbiol. 2022 May;20(5):315. PMID: 33024307; PMCID: PMC7537588.
- Altiok D, Savci EZ, Özkara B, Alkan K, Namdar DS, Tunçer G, Kilinç BR, Suiçmez E, Çetin G, Ünal S, Dönmüş B, Karagülleoğlu ZY, Uncuoğlu DB, Tekeli C, Mendi HA, Bengi VU, Cengiz Seval G, Kiliç P, Güneş Altuntaş E, Demir-Dora D. Host variations in SARS-CoV-2 infection. Turk J Biol. 2021 Aug 30;45(4):404-424. doi: 10.3906/biy-2104-67. PMID: 34803443; PMCID: PMC8573834.
- Soleimanpour S, Yaghoubi A. COVID-19 vaccine: where are we now and where should we go? Expert Rev Vaccines. 2021 Jan;20(1):23-44. doi: 10.1080/14760584.2021.1875824. Epub 2021 Feb 17. PMID: 33435774; PMCID: PMC7898300.
- Dong M, Zhang J, Ma X, Tan J, Chen L, Liu S, Xin Y, Zhuang L. ACE2, TMPRSS2 distribution and extrapulmonary organ injury in patients with COVID-19. Biomed Pharmacother. 2020 Nov;131:110678. doi: 10.1016/j.biopha.2020.110678. Epub 2020 Aug 24. PMID: 32861070; PMCID: PMC7444942.
- Gomes CP, Fernandes DE, Casimiro F, da Mata GF, Passos MT, Varela P, Mastroianni-Kirsztajn G, Pesquero JB. Cathepsin L in COVID-19: From Pharmacological Evidences to Genetics. Front Cell Infect Microbiol. 2020 Dec 8;10:589505. doi: 10.3389/fcimb.2020.589505. PMID: 33364201; PMCID: PMC7753008.
- Glotov OS, Chernov AN, Scherbak SG, Baranov VS. Genetic Risk Factors for the Development of COVID-19 Coronavirus Infection. Russ J Genet. 2021;57(8):878-892. doi: 10.1134/S1022795421080056. Epub 2021 Aug 30. PMID: 34483599; PMCID: PMC8404752.
- Lanjanian H, Moazzam-Jazi M, Hedayati M, Akbarzadeh M, Guity K, Sedaghati-Khayat B, Azizi F, Daneshpour MS. SARS-CoV-2 infection susceptibility influenced by ACE2 genetic polymorphisms: insights from Tehran Cardio-Metabolic Genetic Study. Sci Rep. 2021 Jan 15;11(1):1529. doi: 10.1038/s41598-020-80325-x. PMID: 33452303; PMCID: PMC7810897.
- Zlacká J, Stebelová K, Zeman M, Herichová I. Interactions of renin-angiotensin system and COVID-19: the importance of daily rhythms in ACE2, ADAM17 and TMPRSS2 expression. Physiol Res. 2021 Dec 16;70(S2):S177-S194. doi: 10.33549/physiolres.934754. PMID: 34913351; PMCID: PMC8884363.
- Katopodis P, Randeva HS, Spandidos DA, Saravi S, Kyrou I, Karteris E. Host cell entry mediators implicated in the cellular tropism of SARS‑CoV‑2, the pathophysiology of COVID‑19 and the identification of microRNAs that can modulate the expression of these mediators (Review). Int J Mol Med. 2022 Feb;49(2):20. doi: 10.3892/ijmm.2021.5075. Epub 2021 Dec 22. PMID: 34935057; PMCID: PMC8722767.
- Hubacek JA. Effects of selected inherited factors on susceptibility to SARS-CoV-2 infection and COVID-19 progression. Physiol Res. 2021 Dec 16;70(S2):S125-S134. doi: 10.33549/physiolres.934730. PMID: 34913347; PMCID: PMC8884368.
- Parmar MS. TMPRSS2: An Equally Important Protease as ACE2 in the Pathogenicity of SARS-CoV-2 Infection. Mayo Clin Proc. 2021 Nov;96(11):2748-2752. doi: 10.1016/j.mayocp.2021.07.005. Epub 2021 Jul 15. PMID: 34736607; PMCID: PMC8279956.
- Takeda M. Proteolytic activation of SARS-CoV-2 spike protein. Microbiol Immunol. 2022 Jan;66(1):15-23. doi: 10.1111/1348-0421.12945. Epub 2021 Oct 12. PMID: 34561887; PMCID: PMC8652499.
- Kadam SB, Sukhramani GS, Bishnoi P, Pable AA, Barvkar VT. SARS-CoV-2, the pandemic coronavirus: Molecular and structural insights. J Basic Microbiol. 2021 Mar;61(3):180-202. doi: 10.1002/jobm.202000537. Epub 2021 Jan 18. PMID: 33460172; PMCID: PMC8013332.
- V'kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021 Mar;19(3):155-170. doi: 10.1038/s41579-020-00468-6. Epub 2020 Oct 28. PMID: 33116300; PMCID: PMC7592455.
- Velusamy P, Kiruba K, Su CH, Arun V, Anbu P, Gopinath SCB, Vaseeharan B. SARS-CoV-2 spike protein: Site-specific breakpoints for the development of COVID-19 vaccines. J King Saud Univ Sci. 2021 Dec;33(8):101648. doi: 10.1016/j.jksus.2021.101648. Epub 2021 Oct 19. PMID: 34690467; PMCID: PMC8523302.
- Morgun AV, Salmin VV, Boytsova EB, Lopatina OL, Salmina AB. Molecular Mechanisms of Proteins - Targets for SARS-CoV-2 (Review). Sovrem Tekhnologii Med. 2021;12(6):98-108. doi: 10.17691/stm2020.12.6.11. Epub 2020 Dec 28. PMID: 34796023; PMCID: PMC8596231.
- Peng J, Sun J, Zhao J, Deng X, Guo F, Chen L. Age and gender differences in ACE2 and TMPRSS2 expressions in oral epithelial cells. J Transl Med. 2021 Aug 19;19(1):358. doi: 10.1186/s12967-021-03037-4. PMID: 34412632; PMCID: PMC8374411.
- Viveiros A, Gheblawi M, Aujla PK, Sosnowski DK, Seubert JM, Kassiri Z, Oudit GY. Sex- and age-specific regulation of ACE2: Insights into severe COVID-19 susceptibility. J Mol Cell Cardiol. 2022 Mar;164:13-16. doi: 10.1016/j.yjmcc.2021.11.003. Epub 2021 Nov 11. PMID: 34774871; PMCID: PMC8582230.
- Xu F, Gao J, Bergmann S, Sims AC, Ashbrook DG, Baric RS, Cui Y, Jonsson CB, Li K, Williams RW, Schughart K, Lu L. Genetic Dissection of the Regulatory Mechanisms of Ace2 in the Infected Mouse Lung. Front Immunol. 2021 Jan 8;11:607314. doi: 10.3389/fimmu.2020.607314. PMID: 33488611; PMCID: PMC7819859.
- Clarke SA, Abbara A, Dhillo WS. Impact of COVID-19 on the Endocrine System: A Mini-review. Endocrinology. 2022 Jan 1;163(1):bqab203. doi: 10.1210/endocr/bqab203. PMID: 34543404; PMCID: PMC8500009.
- Abdi A, AlOtaiby S, Badarin FA, Khraibi A, Hamdan H, Nader M. Interaction of SARS-CoV-2 with cardiomyocytes: Insight into the underlying molecular mechanisms of cardiac injury and pharmacotherapy. Biomed Pharmacother. 2022 Feb;146:112518. doi: 10.1016/j.biopha.2021.112518. Epub 2021 Dec 9. PMID: 34906770; PMCID: PMC8654598.
- Deng H, Yan X, Yuan L. Human genetic basis of coronavirus disease 2019. Signal Transduct Target Ther. 2021 Sep 20;6(1):344. doi: 10.1038/s41392-021-00736-8. PMID: 34545062; PMCID: PMC8450706.
- Agolli A, Yukselen Z, Agolli O, Patel MH, Bhatt KP, Concepcion L, Halpern J, Alvi S, Abreu R. SARS-CoV-2 effect on male infertility and its possible pathophysiological mechanisms. Discoveries (Craiova). 2021 Jun 30;9(2):e131. doi: 10.15190/d.2021.10. PMID: 34816001; PMCID: PMC8605861.
- Ragia G, Manolopoulos VG. Assessing COVID-19 susceptibility through analysis of the genetic and epigenetic diversity of ACE2-mediated SARS-CoV-2 entry. Pharmacogenomics. 2020;21(18):1311–1329.
- Hasan MR, Ahmad MN, Dargham SR, Zayed H, Al Hashemi A, Ngwabi N, Perez Lopez A, Dobson S, Abu Raddad LJ, Tang P. Nasopharyngeal Expression of Angiotensin-Converting Enzyme 2 and Transmembrane Serine Protease 2 in Children within SARS-CoV-2-Infected Family Clusters. Microbiol Spectr. 2021 Dec 22;9(3):e0078321. doi: 10.1128/Spectrum.00783-21. Epub 2021 Nov 3. PMID: 34730438; PMCID: PMC8567246.
- Hörnich BF, Großkopf AK, Schlagowski S, Tenbusch M, Kleine-Weber H, Neipel F, Stahl-Hennig C, Hahn AS. SARS-CoV-2 and SARS-CoV Spike-Mediated Cell-Cell Fusion Differ in Their Requirements for Receptor Expression and Proteolytic Activation. J Virol. 2021 Apr 12;95(9):e00002-21. doi: 10.1128/JVI.00002-21. PMID: 33608407; PMCID: PMC8104116.
- Hossain MS, Tonmoy MIQ, Fariha A, Islam MS, Roy AS, Islam MN, Kar K, Alam MR, Rahaman MM. Prediction of the Effects of Variants and Differential Expression of Key Host Genes ACE2, TMPRSS2, and FURIN in SARS-CoV-2 Pathogenesis: An In Silico Approach. Bioinform Biol Insights. 2021 Oct 26;15:11779322211054684. doi: 10.1177/11779322211054684. PMID: 34720581; PMCID: PMC8554545.
- Tarek M, Abdelzaher H, Kobeissy F, El-Fawal HAN, Salama MM, Abdelnaser A. Bioinformatics Analysis of Allele Frequencies and Expression Patterns of ACE2, TMPRSS2 and FURIN in Different Populations and Susceptibility to SARS-CoV-2. Genes (Basel). 2021 Jul 5;12(7):1041. doi: 10.3390/genes12071041. PMID: 34356057; PMCID: PMC8303858.
- Ravaioli S, Tebaldi M, Fonzi E, Angeli D, Mazza M, Nicolini F, Lucchesi A, Fanini F, Pirini F, Tumedei MM, Cerchione C, Viale P, Sambri V, Martinelli G, Bravaccini S. ACE2 and TMPRSS2 Potential Involvement in Genetic Susceptibility to SARS-COV-2 in Cancer Patients. Cell Transplant. 2020 Jan-Dec;29:963689720968749. doi: 10.1177/0963689720968749. PMID: 33108902; PMCID: PMC7593730.
- Jeon S, Blazyte A, Yoon C, Ryu H, Jeon Y, Bhak Y, Bolser D, Manica A, Shin ES, Cho YS, Kim BC, Ryoo N, Choi H, Bhak J. Regional TMPRSS2 V197M Allele Frequencies Are Correlated with COVID-19 Case Fatality Rates. Mol Cells. 2021 Sep 30;44(9):680-687. doi: 10.14348/molcells.2021.2249. PMID: 34588322; PMCID: PMC8490206.
- Lee D. The impact of COVID-19 on human reproduction and directions for fertility treatment during the pandemic. Clin Exp Reprod Med. 2021 Dec;48(4):273-282. doi: 10.5653/cerm.2021.04504. Epub 2021 Nov 26. PMID: 34875734; PMCID: PMC8651760.
- Nayak B, Lal G, Kumar S, Das CJ, Saraya A, Shalimar. Host Response to SARS-CoV2 and Emerging Variants in Pre-Existing Liver and Gastrointestinal Diseases. Front Cell Infect Microbiol. 2021 Oct 25;11:753249. doi: 10.3389/fcimb.2021.753249. PMID: 34760721; PMCID: PMC8573081.
- Generali M, Kehl D, Wanner D, Okoniewski MJ, Hoerstrup SP, Cinelli P. Heterogeneous expression of ACE2 and TMPRRS2 in mesenchymal stromal cells. J Cell Mol Med. 2022 Jan;26(1):228-234. doi: 10.1111/jcmm.17048. Epub 2021 Nov 24. PMID: 34821008; PMCID: PMC8742235.
- Huang Z, Do DV, Beh D, Lee CK, Yan B, Foo R, Tambyah PA. Effects of acute severe acute respiratory syndrome coronavirus 2 infection on male hormone profile, ACE2 and TMPRSS2 expression, and potential for transmission of severe acute respiratory syndrome coronavirus 2 in semen of Asian men. F S Sci. 2022 Feb;3(1):29-34. doi: 10.1016/j.xfss.2021.11.003. Epub 2021 Nov 20. PMID: 34841282; PMCID: PMC8604798.
- Jeong M, Ocwieja KE, Han D, Wackym PA, Zhang Y, Brown A, Moncada C, Vambutas A, Kanne T, Crain R, Siegel N, Leger V, Santos F, Welling DB, Gehrke L, Stankovic KM. Direct SARS-CoV-2 infection of the human inner ear may underlie COVID-19-associated audiovestibular dysfunction. Commun Med (Lond). 2021;1(1):44. doi: 10.1038/s43856-021-00044-w. Epub 2021 Oct 29. PMID: 34870285; PMCID: PMC8633908.
- Sapkota D, Sharma S, Søland TM, Braz-Silva PH, Teh MT. Expression profile of SARS-CoV-2 cellular entry proteins in normal oral mucosa and oral squamous cell carcinoma. Clin Exp Dent Res. 2022 Feb;8(1):117-122. doi: 10.1002/cre2.510. Epub 2021 Nov 2. PMID: 34726347; PMCID: PMC8653086.