More Information
Submitted: June 08, 2021 | Approved: July 03, 2021 | Published: July 05, 2021
How to cite this article: Khan AA, Dutta T, Mondal P, Mandal M, Chowdhury SK, et al. Novel Coronavirus Disease (COVID-19): An extensive study on evolution, global health, drug targets and vaccines. Int J Clin Virol. 2021; 5: 054-069.
DOI: 10.29328/journal.ijcv.1001036
Copyright License: © 2021 Khan AA, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Novel Coronaviruses (nCoVs); World Health Organization; MERS-CoV; SARS-CoV; RNA-dependent RNA polymerase
Novel Coronavirus Disease (COVID-19): An extensive study on evolution, global health, drug targets and vaccines
Abdul Ashik Khan1, Tanmoy Dutta2, Palas Mondal3, Manab Mandal4, Swapan Kumar Chowdhury5, Minhajuddin Ahmed6, Nabajyoti Baildya7, Sourav Mazumdar8 and Narendra Nath Ghosh9*
1Department of Chemistry, Darjeeling Government College, Darjeeling 734101, India
2Departments of Chemistry, JIS College of Engineering, Kalyani-741235, India
3Department of Botany, University of Gour Banga, Malda – 732 103, W.B., India
4Department of Botany, Dukhulal Nibaran Chandra College, Suti-742201, India
5Department of Botany, Balurghat College, Balurghat, West Bengal 733101, India
6Department of Management, Himalayan Garhwal University, Uttrakhand-246169, India
7Department of Chemistry, University of Kalyani, Kalyani-741235, India
8Department of Physics, Dukhulal Nibaran Chandra College, Suti-742201, India
9Department of Chemistry, University of Gour Banga, Malda-732103, India
*Address for Correspondence: Narendra Nath Ghosh, Department of Chemistry, University of Gour Banga, Malda -732103, India, Tel: 09126667601; Email: [email protected]
The Coronavirus disease-2019 (COVID-19), has become a worldwide pandemic and the scientific communities are struggling to find out the ultimate treatment strategies against this lethal virus, Severe Acute Respiratory Syndrome Coronavirus–2 (SARS-CoV-2). Presently, there is no potential chemically proven antiviral therapy available in the market which can effectively combat the infection caused by this deadly virus. Few vaccines are already developed but it is not clear to the scientific community how much efficient they are to combat SARS-CoV-2. Mode of transmission and symptoms of the disease are two important factors in this regard. Rapid diagnosis of the COVID-19 is very much important to stop its spreading. In this scenario, a complete study starting from symptoms of the disease to vaccine development including various SARS-CoV-2 detection techniques is very much required. In this review article, we have made a partial analysis on the origin, virology, global health, detection techniques, replication pathways, doses, mode of actions of probable drugs, and vaccine development for SARS-CoV-2.
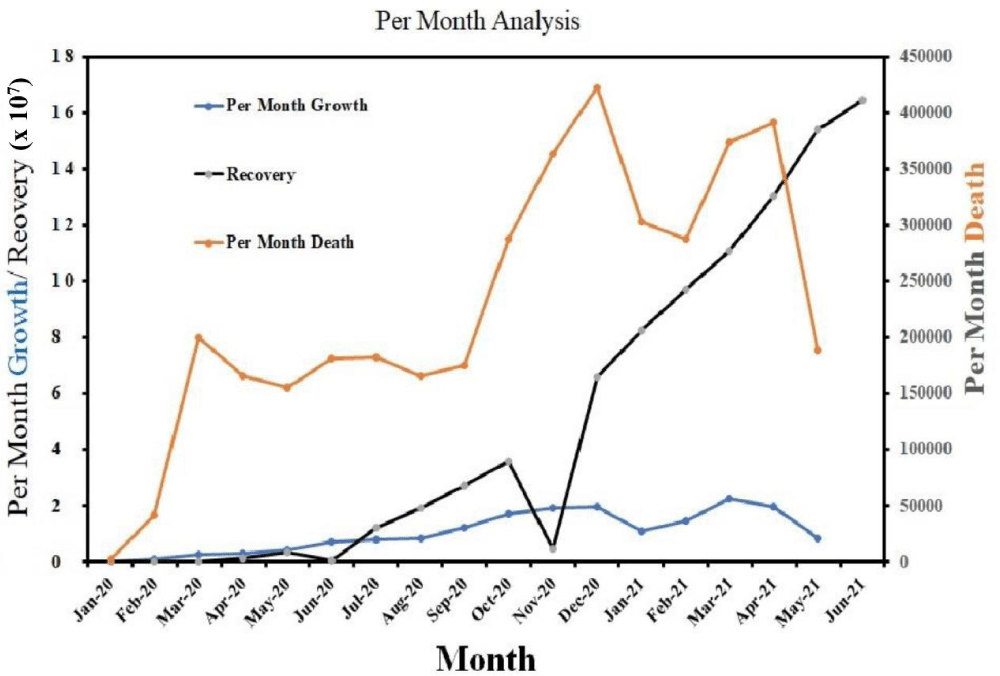
On 31st Dec 2019, a bunch of cases for some unknown types of pneumonia were reported by the hospitals of Wuhan, Hubei, China, knocking the world with an indication of imminent danger [1]. On 1st January 2020, Wuhan public health care authorities closed down the Huanan seafood wholesale market, where live wild animals were sold, due to a suspected link with that outbreak [2]. At the same time, scientists confirmed the 2019 novel coronavirus (2019-nCoV, rechristened as SARS-CoV-2) from the infected pneumonia patients on 7th January, 2020 [3]. On 30th January, 2020, the World Health Organization (WHO) confirmed COVID-19, to be a Public Health Emergency of International Concern (PHEIC), and declared as an epidemic [3]. As on 7th June, 2021, globally 173,937,118 affected and 3,741,055 death cases were confirmed by the WHO. Similar symptoms like, common cold, fever in human beings, caused by Coronavirus, were reported in the middle of the year 1960 [4]. Coronavirus (CoVs), a genus of the Coronaviridae family, is likely one of the major pathogens by which the human respiratory system is affected. Two classes of CoVs were first isolated i.e. HCoV-229E and HCoV-OC43 [5]. The HCoV-229E straining was related to symptoms of common cold [6]. An outbreak also occurred in the Guangdong province of southern China, caused by a similar pathogen SARS-CoV in 2002, where approximately 38% of all patients suffered from acute breathing problems and hence required artificial respirators [7,8]. The overall mortality rate of SARS-CoV in 2002 was approximately 10%, where older people accounted for approximately 50% of the cases, while middle aged and children comprised the other 50% [9]. In the September 2012, another viral infection occurred in Saudi Arabia which subsequently spread to the Middle East countries like Qatar, Germany, Jordan, France and Britain and it was named as Middle East Respiratory Syndrome (MERS) [10]. Symptoms were almost similar to the previous one (SARS-CoV-2002) like fever, diarrhea, cough and breathing problem [11]. Recently, SARS-CoV zoonotic pathogens have mutated to form themselves a new virus “2019 novel Coronavirus” (2019-nCoV) or SARS-CoV-2 formally named by the World Committee on Virus Classification. Erstwhile, 2019-nCoV was also known as “Novel Coronavirus Pneumonia (NCP)” in China till 8th February 2020 and subsequently has spread abruptly to all over the world [12-14]. It is explained by the research group of Ji, et al. in their investigation that the codon usage of 2019-nCoV is very similar to the host, snakes than any additional hosts for cross-species transmission of the pathogens to human [15]. The transmission of this respiratory pathogens is associated with three main primary modes e.g. “airborne”, “contact,” and “droplet” transmission but the research-based literature and the guidance from the public health agencies are still paradoxical [16]. As per the research based studies, physical touch is mainly responsible for the contact transmission and indirectly can transmit by settled droplets [16]. COVID-19 spreads by human-to-human close contact [17,18]. The symptoms of COVID-19 infection may appear in 5-14 days [19]. The time duration from the onset of COVID-19 symptoms to eventual death of the patients ranges between 6 to 41 days [20]. This period varies from patient to patient depending on their age and immune response of the system. The period is shorter for patients above 70 years old compared to those less than 70 years old [20]. The general symptoms of COVID-19 are fever, fatigue, cough, while additional symptoms like diarrhea, sputum production, headache, lymphopenia, haemoptysis, dyspneamay manifest, varying from patient to patient [17,21]. COVID-19 infection showed a few unique clinical features like targeting the lower airway which causes the upper respiratory tract infection like a runny nose, a sore throat and sneezing [22]. It has been observed in per month analysis that the worldwide growth rate of infection and the death rate of COVID-19 patients has increased aggressively whereas the recovery rate of the patients from this disease is quite low with such rates (Figure 1). CoVs are transmitted as SARS-CoV and MERS-CoV to alternate host civet cats and camels respectively, prior to affecting the humans, but presently, there is no clear evidence of the origin of SARS-CoV-2 (2019-nCOV) [23,24].
Figure 1: Per month new COVID-19 cases trend from January 2020 to June 2021the diagram represents that the growth and death rates are exponentially increasing (Data source: https://www.worldometers.info/coronavirus/?fbclid=IwAR35ZFiRZJ8tyBCwazX2N-k7yJjZOLDQiZSA_MsJAfdK74s8f2a_Dgx4iVk;Date: 24.06.2021).
According to the WHO, there are no recommended medicines available in the market and hence maintaining social distance, along with proper hygiene, is the only way to stop the spread of COVID-19 [25]. Though there are a few myths regarding the medicine of COVID-19 (mixture of different kinds of herbals) but it is not approved by the FDA [25,26]. Since January 2020, at least 37 bio-pharmaceutical companies worldwide are involved for the development of the drug and protective vaccine with multiple platforms including adenoviral vector, mRNA, recombinant protein and DNA [27]. Researchers are also trying to use some natural compounds against this virus [28-30]. Some drugs which are under investigation viz. Hydroxychloroquine, Carfilzombic, Eravacycline, Valrubicin, Lopinavir needs to be successful during in-vivo analysis [28,31-36].
The objective of this review article is to summarize all the aspects of COVID-19, starting from its history to the present scenario and develop a clear understanding of the transmission, detection, precaution, symptoms, and virology along with the ways of treatment of COVID-19 via probable drugs.
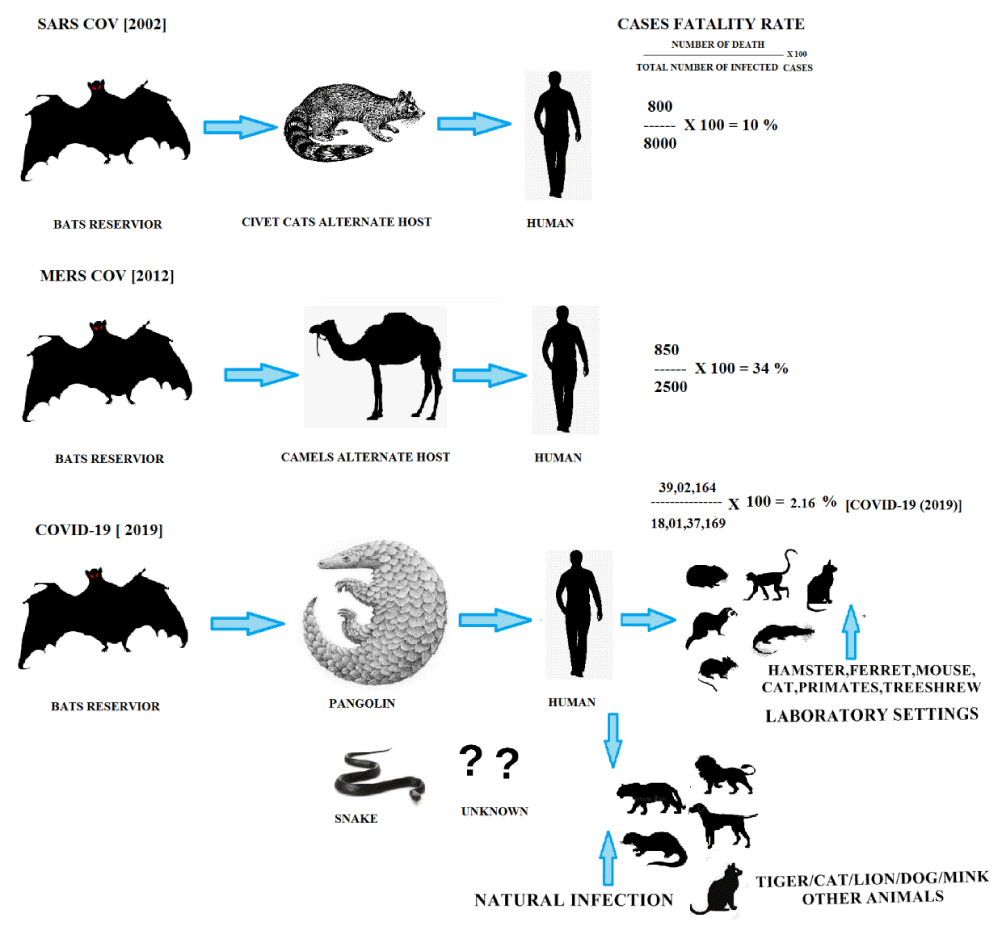
Previously, the CoVs like SARS-CoV and MERS-CoV pathogens were transmitted from different animals including bats, cats, cattle and camels to the human body as shown in figure 2. Over the years, much has been learned from the global outbreak pattern for similar pathogens occurring in China, South Africa, Saudi Arabia (Middle East Countries) and the pandemic spread, affected from severe respiratory diseases.
The recent outbreak of 2019-nCoV, its origin and its transmission to the human body is under debate. However, from the epidemiologic investigation at the starting point of this global outbreak, it was reported that 2019-nCoV infected patients were seafood vendors, visitors or were close to the infection at the seafood market of Wuhan [37]. Till now the detailed host source of SARS-CoV-2 and its transmission to human body is not clear (Figure 2). SARS-CoV is transmitted by respiratory droplets (coughs or sneezes), resembling the spread of influenza from infected persons [38]. It may also spread through an airborne route at a close distance, eg., person-to-person [39]. The transmission rate (R0) (reproductive number or the usual number of persons to that a single infected person shall spread the pathogens) of COVID-19 initially is between 1.4 to 2.5 projected by the WHO and higher values of R0 varies in the range of 3.6 to 4.0 reported by different literature [40]. Accordingly, these values would be associated to the R0 of 1.3 for common flu and R0 of 2.0 for SARS-CoV. If the R0 value is not more than 1, the virus may disappear, else the epidemic shall remain in the environment [41,42].
Figure 2: Modes of transmission of CoVs.
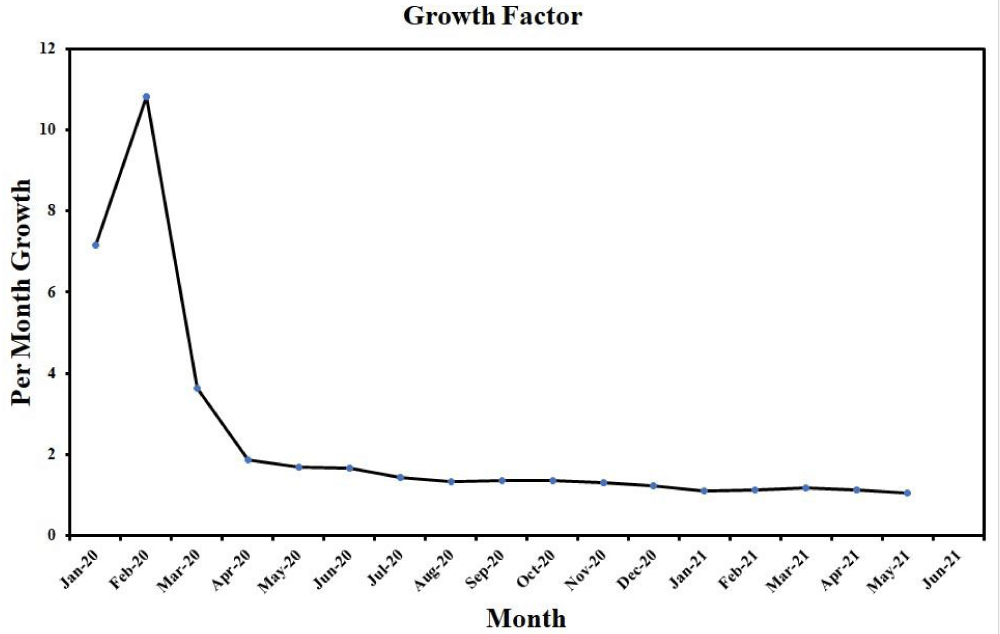
The 2019-nCoV infection is very contagious, known to all that the virus can spread rapidly from one person to other and in many instances the virus can spread even prior to becoming symptomatic [1].The growth factor of COVID-19 cases over the globe is shown in figure 3. Growth factor related to the number of months where a quantity reproduces itself over time. The formula used here is: every day’s new cases divided by fresh instances on the preceding day.
Figure 3: Growth factor curve of COVID-19 from January 2020 to June 2021. (Data source: https://www.worldometers.info/coronavirus/?fbclid=IwAR35ZFiRZJ8tyBCwazX2N-k7yJjZOLDQiZSA_MsJAfdK74s8f2a_Dgx4iVk ; Date: 24.06.2021).
The virus comes in the bronchial secretions with a droplet transmission, where one having infectious cough, sneezing can infect another in close contact, directly through the mucous membranes. Infections to an otherwise healthy individual can also spread by facial touch (touching one’s mouth, nose, and eyes by their hands, which are already exposed to the infection). 2019-nCoV persisted in the aerosols during experiments at least three hours as described in a letter of an editor [43].
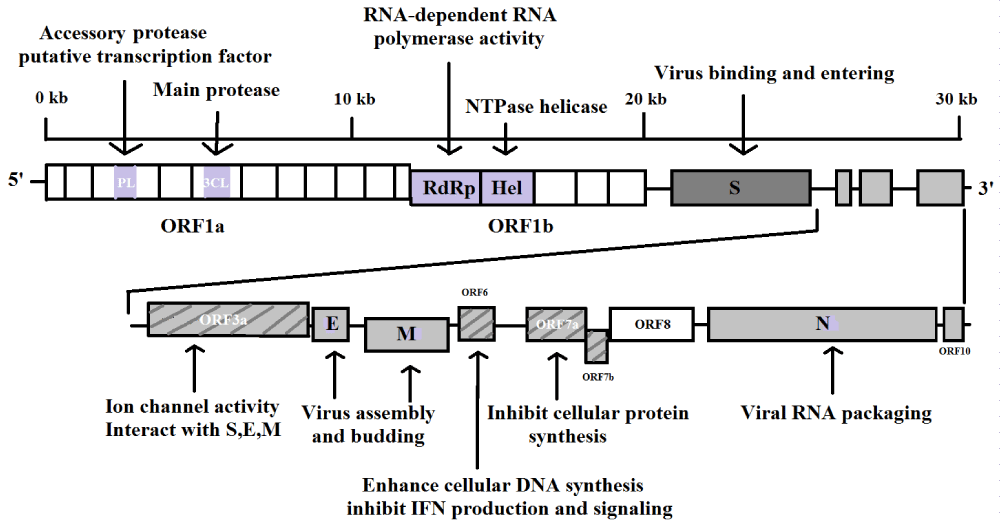
The structure of SARS-CoV-2 is composed of four different types of proteins viz. spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins. According to the cryo-electron microscopy analysis, coronavirus genome is a large, linear single stranded, positive-sense RNA molecule that contains all information necessary for the making of viral replication. In positive sense, the size of the SARS-CoV depends on its proteins (S, E, M, N) and the other helper proteins and it varies from 29.0 kb to 30.2 kb and the molecular weight is usually 5.5 x 106 to 6.1x 106. There are total seven genes of CoVs RNA genome that are preserved in the following order: ORF1a, ORF1b, S, OEF3, E, M, N in 5′ to 3′ direction. Major part of the RNA genome is coated by ORF and rest of the part of the genome shield by mRNA and other necessary proteins. ORF1a/b develops replicase proteins PP1a and PP1ab through ribosomal frame shifting and simultaneously both these proteins produce sixteen non-structural proteins (NSPs). These NSPs also carry on the process of various viral activities including the reproduction of the replicase transcriptase compound. The mRNA part is responsible for the formation of structural proteins (S,E,M,N) and other accessory proteins[44] as shown in figure 4.
Figure 4: Genome of SARS-CoV-2.
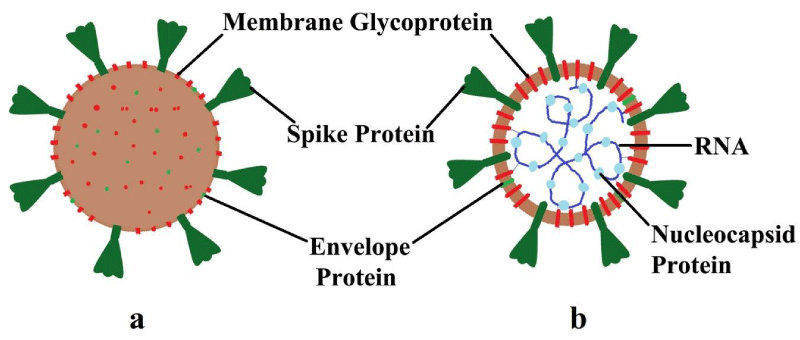
Furthermore, there is a protein envelope named HE protein and the RNA packed nucleocapsid protected by the protein embedded lipid layers [45,46]. The outer and inner portion of SARS-CoV-2 is shown in figure 5.
Figure 5: Structure of SARS-CoV-2 (a) Outside; (b) Inside.
The nature and the structure of the RNA genome of SARS-CoV-2, is very similar to that of the family of beta-CoV, which occurred as pandemic previously. However, the contamination of SARS-CoV-2, spreads rapidly through the droplets and person to person and the integrity of the envelope is crucial for viral infection but the lipid layer enveloped of SARS-CoV-2 can be easily destroyed by lipid solvents such as detergents, alcohol and some another disinfectants [47-49].
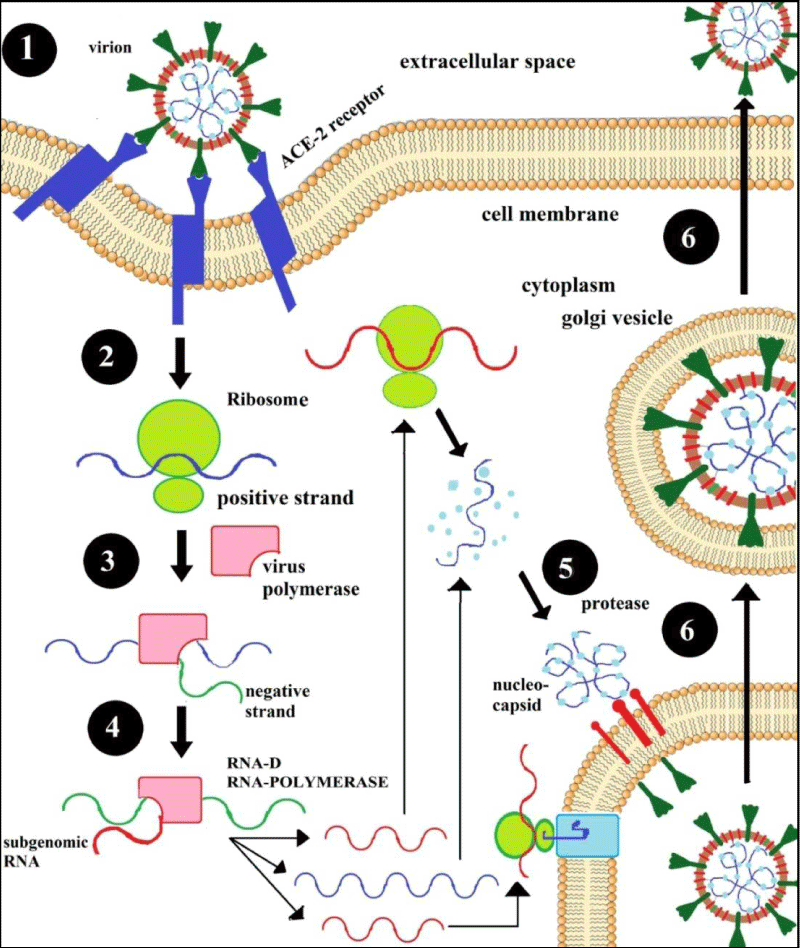
To infect a host cell, first the spike glycoprotein (S) of corona virus must bind the host cell surface with the help of a receptor called angiotensin converting enzyme‑2 (ACE‑2) [50] and the conformational changes occurring that allows the virus to get inside by the process known as endocytosis. After that virus genome enters into the host cell cytoplasm and release nucleocapsid or ssRNA(+) followed by replication transcription complex [51]. Translation process is carried out by involvement of produced pp1a and pp1b from the ribosomal shifting of viral genome ORF1a and ORF1b and producing dsRNA genome from the genomic ssRNA(+) [51,52]. The dsRNA genome is transcribed or replicated and providing viral mRNAs or new ssRNA(+) genomes by the 16NSPs. Total 16 NSPs which are produced from PPs involving in specific functions mainly for the replication process such as suppression of host cell by NSP1 and NSP2. The production of multi-domain complex by NSP3, replication by M protease (NSP5), [52] transmembrane (TM) proteins (NSP4 & NSP6), NSP7 & NSP8 acting as primase [53] and R-binding protein NSP9 is efficient for viral infection in the dimeric mode [54,55]. NSP10 acts as co-catalyst at the beginning of replicative enzyme and NSP11 & NSP15 shows endo-ribonuclease activity [56,57]. NSP12, NSP13, NS14 and NSP16 showed RNA‑dependent RNA polymerase activity, helicase activity, exoribonuclease activity and methyl transferase activity respectively [53]. Synthesis of structural proteins encoded by subgenomic mRNAs, the central organiser of CoV assembly actually determine the shape of viral envelope after synthesis of proteins such as M, E and S by entering into the endoplasmic reticulum (ER) i.e. Golgi intermediate compartment (ERGIC) complex [58]. At the same time, ribonucleoprotein is formed by the complexion of replicated genome and N protein and new virions are released by exocytosis from ERGIC [59,60]. The step by step translation and replication process of CoV in the host cell is depicted in figure 6.
Figure 6: Replication pathway of SARS-CoV-2 in the host cells.
The extremely non-specific symptoms of the COVID-19 include fever, respiratory symptoms like- cough, viral pneumonia and dyspnea [61]. During the whole course of disease COVID-19 different patients came across different problems like gastrointestinal disorders, diarrhea, and respiratory problems [62]. In most of the cases, the delay in detection at early stage is the main reason of the high spread of this virus. Recent reports on the Diamond Princess Cruise ship quarantined in Yokohama, Japan, showed that 454 infected cases identified out of 3700 passengers and crew, as they did not have enough diagnosis kits [63]. Rapid screening and early detection of SARS-CoV-2 in suspected patients has become necessary to control the lethal effect of the COVID-19 outbreak [64,65]. The Point of Care (POC) device, which is very efficient and cost-effective will be helpful for early stage detection [66]. Some modern detection techniques are summarized as follows.
Polymerase Chain Reaction (PCR) assay for COVID-19
Real time reverse transcription polymerase chain reaction (rRT-PCR) is currently used as standard molecular technique for the detection of COVID-19 [67]. Sophisticated laboratory equipment are required for rRT-PCR technique (Bio-safety level 2 or above) [68]. Commercial PCR based techniques are usually time taking and expensive and it requires experts as the existence of RNA pathogen does not always reflect the disease [69-71]. Another problem with Commercial PCR system is that other respiratory viruses are also encountered at the time of detection ofCOVID-19 that reflects positive CoV PCR result [71].
Loop Mediated Isothermal Amplification Assay (LAMP) for COVID-19
It is a new method where nucleic acid (DNA) is amplified with high efficiency, rapidity and specificity in isothermal condition. A set of four designed primers with a DNA polymerase are utilised. By this method the stand displacement activity synthesize the targeted DNA up to 109 copies below 60 °C within one hour [72]. With several reversed replications of the target stem loop DNAs are obtained as final product and appears like a cauliflower. LAMP is very much popular for pathogen detection and it is more simple than PCR [73]. Though LAMP is very much faster and sensitive than PCR but its establishment is in early stage.
CRISPR-based diagnostics
When SARS-CoV-2 was first identified, many scientists redirected their focus to CRISPR (Clustered regularly interspaced short palindromic repeats)-based diagnostics to detect SARS-CoV-2. The use of Cas12/Cas13 enzymes is the key principle of this approach and it is initially determined as the components of ‘bacterial immune system’ coupled to RNAs with specific binding to certain regions of the target DNA or RNA [74]. STOP Covid (SHERLOCK Testing in One Pot COVID) and DNA Endonuclease-Targeted CRISPR Trans Reporter are two commonly used CRISPR-based detection methods for SARS-CoV-2 [75]. The first technique is targeted to the N-gene and E-gene of SARS-CoV-2 and engages the Cas12a enzyme, on the other hand, the second one is targeted at the N gene with the utilization of Cas12b enzyme. PCR-based tests take center stage to maintain the standard of diagnostic tests [76,77]. However, a labor-intensive protocol with long waiting for results, and a dwindling reagents supply has led to many scientists looking for alternative CRISPR test. For mass testing, many have turned to CRISPR test, as its nucleotide-targeting ability makes it optimal for the detection of the presence of viral RNA.
LFIA detection method
Lateral Flow Immunoassay (LFIA) is another conventional method for the detection of COVID-19 disease. After the addition of the sample, specific IgM and IgG antibodies flow with the capillary action and specific immunoglobulins bind with SARS-CoV-2 gold conjugated antigens. The main advantage of this method is that it is suitable to detect COVID-19 disease at the different stages of the infection due to the combined determination of IgM and IgG [77].
Virus-proteins or specific antibodies against these SARS-CoV-2 virus-proteins
It is the modern method for the detection of COVID-19 including antibody and antigen tests. Here the interaction between SARS-CoV-2 virus proteins and specific antibodies is performed. As a result, an intermediate complex is formed which gives pieces of information regarding the kinetics of SCoV2-rN/anti-SCoV2-rN composite formation which gives the SCoV-rN and anti- SCoV-rN Gibbs free energy (DGAssoc). The results are very helpful for the design of new diagnostic systems for the development of new anti-SARS-CoV-2 medicine [78].
The early symptoms of COVID-19 infection are fever, sore throat, tiredness, fatigue, dry cough, aches and pains, pneumonia, dyspnea, swollen eyes and sometimes diarrhea, nausea or a runny nose and sneezing, for a low percentage of people [21,79]. These symptoms are shown within five to six days after the infection transmission of SARS-CoV-2 but the incubation period may last upto 14 days [70,80,81][70,80,81]70, 80, 8169, 79, 8068, 78, 7967, 77, 7867, 77, 7868, 78, 7968, 78, 79. The virus particles spread through the droplet transmitted and largely affect the nasal and mucous membranes in the throat and surroundings, which infect the neighbouring cells. They crawl progressively down and affect the bronchial tubes mucous membranes and lungs. They can damage the alveoli or the lung sacs, and the lung function, since the lung is filled with fluid, pus and dead cells. Patients suffering from excessive breathing trouble may need ventilation support (Acute Respiratory Distress Syndrome). Extreme condition may results death [82][82]82818079798080.
Sometime it may affect the rectum of the infected individual. It may enter the cells of gastrointestinal system hence causing symptoms like diarrhea or indigestion. The pathogen may directly damage the bloodstream, blood vessels, bone marrow and organs like the liver, the heart, the kidney. This may result in inflammation and malfunction of all the organs. Some patients may tolerate the damage of organs and the infection with immune system. It is not reported that 2019-nCoV could not damage the brain but could insinuate the brain in some patients. The 2019-nCoV may infect some nerve cells also [39].
COVID-19 is a respiratory disease by which people are being affected with minor to modest symptoms and may get cured without any specific treatment. Most of the patients infected with SARS-CoV-2 take 1 to 14 days to grow symptoms [83]. The SARS-CoV-2 can infect individual in different ways. It is observed that the people whose aged are more than 60 years, with pre-existing diseases, may have higher risk of getting affected. The symptoms of COVID-19 may appear after exposure as MERS-COV virus and incubations period of SARS-COV-2 is approximately 5.2 days [84].
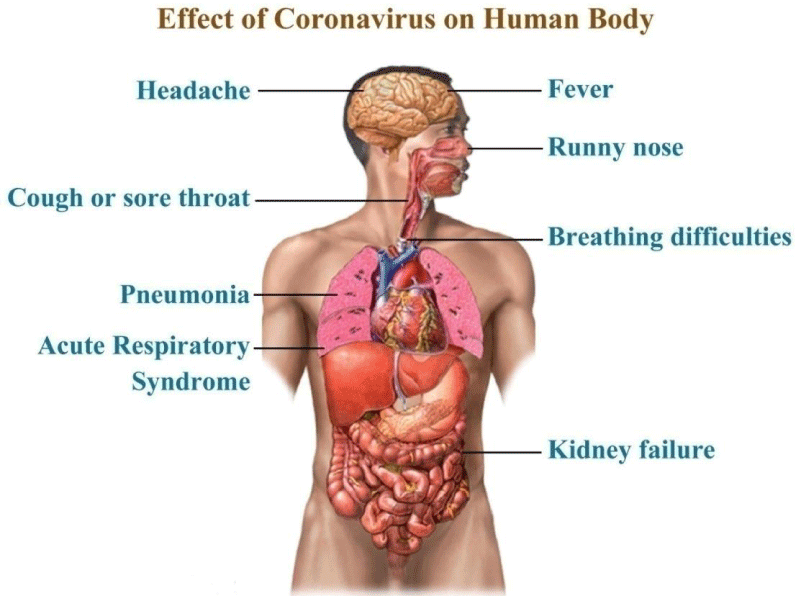
Emergency warning signs of COVID-19 such as acute breathing trouble, tremendous chest pain or pressure, new confusion or inability to arouse, bluish lips or face, if developed, required immediately medication. The situation report - 95 from the World Health Organization reveals 84570 and 1716078 confirmed cases in China and USA respectively as on 31st May, 2020. The effect of corona virus on human body is shown in figure 7.
Figure 7: Symptoms and lethal effects of CoVs in the human body.
Individual exhibiting COVID-19 symptoms must undertake a few necessary precautions to prevent the human-to-human transmission. The expert recommendations related to precaution for avoiding COVID-19, by the WHO and the CDC are i) Anyone who has COVID-19 symptoms, must stay at home and avoid large or any mass gathering, occasions, or programmes & public areas. Avoid public transportation, taxis or ride-sharing, and maintain distance (about 6 feet) from others. Keep away from affected area or community with COVID-19 cases. ii) Wash hands frequently with soap-water for more than 20 seconds or use hand sanitizer containing alcohol (not less than 60%) and cover mouth & nose with a tissue or the elbow while sneezing or coughing. Used tissue must be thrown into covered dustbin or lined trashcan. Also stop touching mouth, nose and eyes with unless hand sanitize. iii) Wash and disinfect surfaces frequently on regular basis, do not take raw or undercooked meat, and do not share household items like glasses, cups, dishes, eating utensils, bedding and towels etc. Use separate bathroom if possible and finally, stay isolated in a particular room (termed home quarantine) away from the other members of family and with limited access to pets. iv) Visit a doctor immediately in case of any COVID-19 symptoms, informed the clinic beforehand so that they can take necessary precautions, and disclosed to the doctor any travel history or contact with contaminated or infected people.
The authorities of the health departments of different countries have appealed to and instructed all concerned regarding epidemic precaution, prevention, protection, treatment, medicine and diagnostic techniques; including awareness and information, education and communication (IEC) activities, for prevention of such severe respiratory diseases due to CoVs [85].
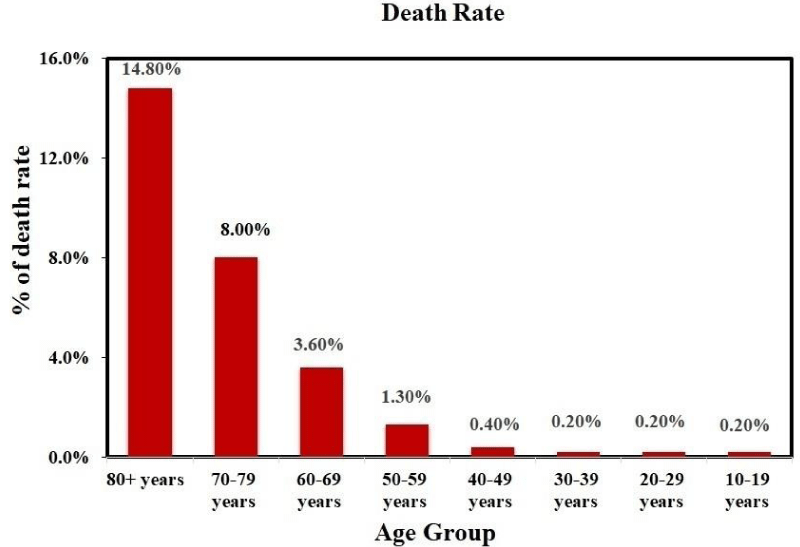
Though COVID-19 can affect any age group, its deadliest effect is mostly found in middle and older age group. Recent investigations reveal that it has a profound effect in the age range of 49 to 56 years [40] and 77.8% of confirmed patient were within the age of between 30 to 79 years among 44672 patients for period of 16th to 20th February 2020 [86]. Recent reports showed that the mortality rate is 0.2% for the 10-39 years age group, 0.4% for 40-49 years age group, 1.30% for 50-59 years age group, 3.6% for 60-69 years age group, 8% for 70-79 years age group and highest rate of 14.8% is realized for 80+ age group as shown in figure 8. Presently, the limited data available pertaining to age-related risk-factors is insufficient in case of COVID-19.
Figure 8: Age dependent effect of COVID-19; (Data source: https://www.worldometers.info/coronavirus/?fbclid=IwAR35ZFiRZJ8tyBCwazX2N-k7yJjZOLDQiZSA_MsJAfdK74s8f2a_Dgx4iVk ; Date: 24.06.2021)
Recent data shows people with aged above 65 years and suffering from chronic bronchial diseases or mild to severe asthma, severe chronic cardiac disease, high obesity (body mass index [BMI] ≥ 40) have a high risk of COVID-19 infection. Furthermore, people with certain uncontrolled diseases like diabetes, chronic kidney diseases, or hepatic diseases, immunity disorder, bone marrow or organ transplantation, HIV or AIDS also have an elevated risk of getting affected by COVID-19 [42,43][42,43]42, 4341, 4240, 4139, 4039, 4040, 4140, 41.
At present vaccines or drugs are not available in the market that can effectively combat the COVID-19 disease [31]. Since COVID-19 is a viral infection, antibiotics readily available are not effective. Different drugs were experimented through in vitro trails on the human-based SARS-CoV and MERS-CoV, but they are initial stages. Interferon-beta (IFNb) has potential to reduce the replication of MERS-CoV only but there are no appropriate drugs available in the market for SARS-CoV-2 [87]. Initially, usage of the drug with the combination of Anti-HIV drugs Lopinavir/Ritonavir (LPV/RTV), ribavirin and pegylated interferon resulted a success report to cure MERS-CoV infected patients in South Korea [40,88]. Furthermore, in vitro studies with the drug remdesivir, a powerful terminator of RNA viral transcription is at beginning stage for the diseases caused by SARS-CoV, MERS-CoV and related zoonotic bat CoVs [89-91]. There are few drugs such as captopril, perindopril, ramipril, lisinopril, benazepril and moexipril also tested for effectiveness toward ACE2 as the similarity found with ACE [92].
Genomic information and replication process of SARS-CoV-2 is almost similar to the SARS-CoV, but the only difference is that it has higher binding affinity to host cell. To discover potential drugs for SARS-CoV-2 we need to analyse systemic replication and transcription pathways of the virus in the host cell. The replication of SARS-CoV-2 begins with the binding of its spike protein (S) on the cell-surface molecules of the host. This receptor recognition is important for initiating virus entry into the host cells, thereby playing a major part in the tissue and host species tropism of viruses. Therefore inhibitory effect of the communication between the spike proteins to the human related proteins specifically enzymes viz. humanACE2 and type-II trans-membrane serine protease (TMPRSS2) enzymes is the key step for drug development. Secondly inhibition of the viral enzyme and proteins that are involved for the synthesis and replication of RNA is important for designing potential drugs. Based on the homology there are total 18 viral proteins (Figure 4) viz. Nsp1, Nsp3 (Nsp3b, Nsp3c, PLpro and Nsp3e), Nsp7-Nsp8 complex, Nsp9–Nsp10 and Nsp14–Nsp16, 3CLpro, E-channel (E-protein), ORF7a, Spike, N-terminal RNA binding domain (NRBD), C-terminal RNA binding domain (CRBD), RdRp, helicase and 2 human enzymes viz. ACE2 and TMPRSSS2 that are involved from the endocytosis to the exocytosis of SARS-CoV-2. Interrupting endocytosis and exocytosis processes would be potential for development and discovery of new drugs therapeutics.
Before going to in details of the drugs related to the SARS-CoV-2, herein we note some approved drugs that are reported for the treatment of SARS-CoV (2002). These are the following drugs: Ribavirin (inhibits viral polymerase) [93], E64-d,glycyrrhizin (inhibits replication process) [94].
Drugs that effectively block the host specific receptor or enzymes
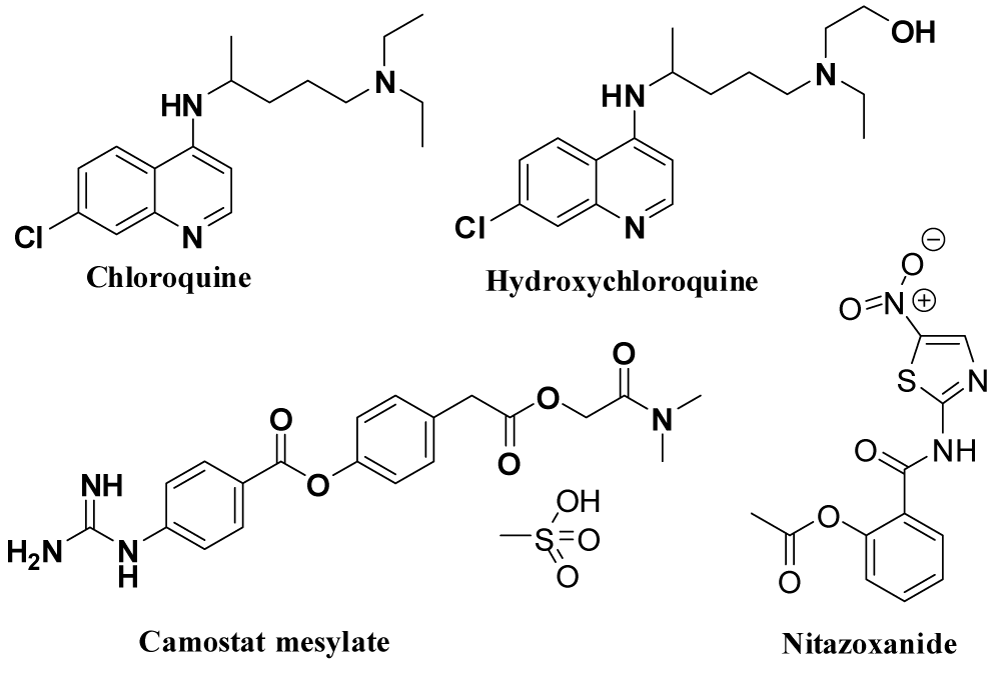
ACE-2 enzyme inhibitors: ACE2 enzyme, a human host spike receptor helps for cross species transmission. The spikes of SARS-CoV-2 have more affinity to ACE2 receptor binding domain (RBD) as compared to previous MERS & SARS CoV. There is a strong interaction between several key amino acid residues of RBD receptor and ACE2 enzyme [95]. The drugs that showed inhibitory activity against ACE-2 enzyme areeprosartan (Tevetan), irbesartan (Avapro), losartan (Cozaar), olmesartan (Benicar), telmisartan (Micardis), valsartan (Diovan) and chloroquine along with their mode of action and side effects are shown in the table 1. Among these drugs only Cholroquine is reported to have potential activity against the interface between the ACE-2 and spike protein [96]. Few ACE-2 enzyme blocker drugs are shown in figure 9.
| Table 1: Drugs that block the host specific receptor or enzymes. | |||
| Sl. No | Drug | Mode of Action | Comments |
| 1. | Chloroquine [96,100,101] | Choroquine is an anti-malarial drug with the properties of anti-inflammatory and immunomodulatory. Its mechanism involves blocking the interface between the spike glycoprotein (S) of virus and the ACE2 at the surface of the cell membrane inhibiting fusion of the virus and immunomodulation of cytokine release. | Irreversible damage to retina, deafness, hearing loss, increased liver enzyme, anorexia, nausea, vomiting, tinnitus. In vitro analysis of chloroquine has potent against SARS-CoV-2 with an EC50 at 48 hours of 1.13 μM in Vero E6 cells. |
| 2. | Hydroxych-loroquine [96,100,101] | Follow the same mechanism of previously mentioned chloroquine like inhibit the catalytic activity of viral enzymes or processes such as viral DNA and RNA polymerase, viral protein glycosylation, virus assembly, new virus particle transport and virus release. | Due to long term use, it may damage retina, dizziness, muscle pain, stomach upset. Risk of cardiac arrhythmias. In vitro analyses in Vero cells demonstrated that the potency of hydroxychloroquine (EC50 of 0.72 μM) was greater than that of chloroquine (EC50 of 5.47 μM) against SARS-CoV-2. |
| 3. | Camostatm-esylate (Foipan) [102,103] | Japanese drug camostatmesylate is a TMPRSS2 inhibitor and also block the cellular entry of SARS-CoV-2. | Under phase 1 trial against SARS-CoV-2. |
| 4. | Nitazoxani-de [104,105] | Nitazoxanide, broad-spectrum antiviral drug inhibits the action of viral protein expression which interferes with host regulated pathways involved in viral replication rather than virus-specific pathways. | Common side effects may include: nausea, stomach pain; headache; or discolored urine. It has demonstrated potent in vitro activity against SARS CoV-2, with an EC50 at 48 hours of 2.12 µM in Vero E6 cells. |
Figure 9: Drugs that effectively block the host specific receptor or enzymes.
Transmembrane protease, serine 2 (TMPRSS2) inhibitors: TMPRSS2 is the serine protease act as Spike (S) protein priming. Aprotinin (Trasylol) is common available drug for a competitive inhibitor of several serine protease [97]. The enzyme activity of TMPRSS2 inhibitor is able to prevent CoVs from host molecules entering [98]. Few anti-bacterial drugs viz. pivampicillin, hetacillin, cefoperazone and clindamycin and anti-virus natural compounds viz. phyllaemblicin G7, neoandrographolide, kouitchenside were already targeted for TMPRSS2 inhibitors [99].
Drug for inhibiting virus structural proteins
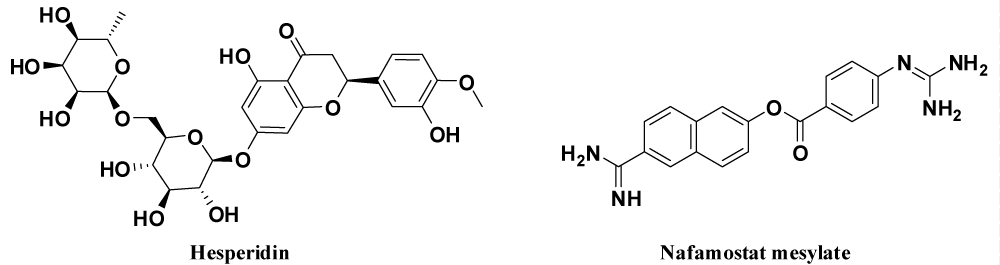
Drugs for spike glyco-protein inhibition: Spike is the structural protein of SARS-CoV-2 that binds to the host cell and sliced into S1 and S2 by the host cell protease. The role of S1 is to bind ACE2, while S2 mediates virus–cell and cell–cell membrane fusion. Therefore, drug target is to bind to the spike and inhibit the entry of CoV. Table 2 contains drugs that inhibit viral structural protein with their mode of action and side effects and figure 10 shows structures of few drugs.
Figure 10: Drugs for inhibiting virus structural proteins.
| Table 2. Drug for inhibiting virus structural proteins. | |||
| Sl. No. | Drug | Mode of Action | Comments |
| 5. | Hesperidin [106] | Hesperidin, flavanone glycoside (found in the citrus including lemons, grapefruits and sweet oranges) was expected to be in middle thin pat of the RBD surface of Spike and the dihydroflavone part of the compound moved similar with the β-6 sheet of RBD. By superimposing the ACE2–RBD complex to the hesperidin–RBD complex, a different overlap of hesperidin with the line of ACE2 could be observed. | Side effects include stomach pain and upset, diarrhea and headache. Considered for clinical trial but no data exist now. |
| 6. | Nafamosta-tmesylate (Fusan) [107,108] | Japanese researchers suggested that Nafamostat can prevent spike-mediated membrane fusion of the virus with the host cell surface proteins, the first step in SARS-CoV-2 infection. | Approved for anticoagulant therapy in Asian countries |
| 7. | REGN3048-3051 [109] | REGN3051 is a fully human monoclonal antibody (mAb) which binds to the spike protein of the MERS-CoV and inhibits the interaction between the virus and host cell. | Currently in phase 1 trial against COVID-19. |
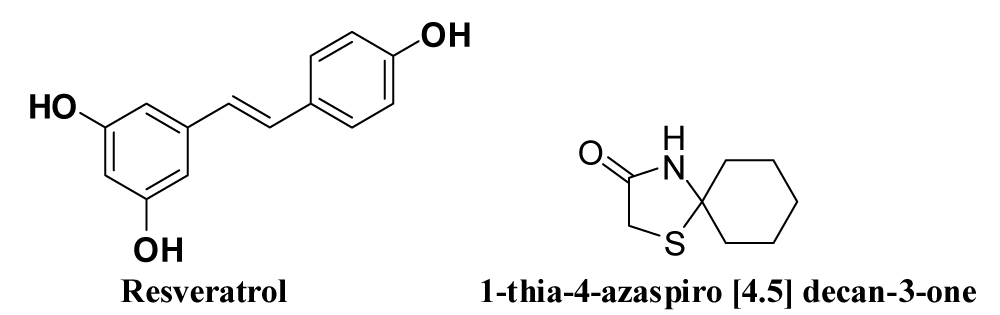
Drugs for E & N protein (E-channel) Inhibition: E protein (E-channel) possesses important biological functions for the structural integrity of coronavirus and host virulence. CRBD and NRBD of CoVs N protein are required for N proteins in host cell to bind with CoVs RNA efficiently. Therefore, inhibition of E protein or N protein (NRBD & CRBD domains) can be used for the discovery of anti-viral drugs. Table 3 lists some drugs that inhibit E & N protein along with their mode of action and side effects and the corresponding structures of these drugs are given in figure 11.
| Table 3: Drugs for inhibiting E &N protein. | |||
| Sl. No. | Drug | Mode of Action | Comments |
| 8. | Resveratrol [110] | Resveratrol, a natural compound that is found in grape seeds and skin and in red wine, inhibits the expression of nucleocapsid (N) protein essential for replication of virus in the host cell. | Resveratrol can be given either at high dosages up to 250 μM or at a relatively low concentration, such as 62.5 μM consecutively to treat MERS-CoV-infected cells. No data exist against COVID-19. |
| 9. | 1-thia-4-azaspiro [4.5] decan-3-one derivatives [111] | It is mainly fusion inhibitors and also inhibits human coronavirus 229E virus replication. | Most potent analog having an EC50 value of 5.5 µM. |
Figure 11: Drugs for inhibiting E & N proteins.
Drugs for inhibiting virus non-structural proteins (NSPs)
SARS-CoV PL main protease inhibitors: Two essential protease inhibitors namely papain-like protease (PLpro) and a 3C-like protease (3 CLpro) intervene the replicase poly-proteins pp1a and pp1b.
i. 3C-like main protease (3CLpro): The 3CLpro (Nsp5) is essential in the life cycle of the virus through the formation of mature enzyme and releasing downstream Nsps (Nsp4–Nsp16). Thereafter, the blockage of 3CLpro is an attractive targeted by the drug therapy for the COVID-19.
ii. Papain-like protease (PLpro) inhibitors: PLpro is an important protease inhibitor for rectifying virus reproduction by the cleavage of N-terminus of the replicase poly-protein to discharge Nsp1, Nsp2 and Nsp3. It also suppresses the innate immunity of the host cell. Therefore, PLpro may be standard target for Coronavirus inhibitors. Table 4 summarizes the protease inhibitor drugs along with their mode of action and their side effects. The corresponding structures of those drugs are shown in figure 12.
| Table 4: Drugs for inhibiting protease inhibitors. | ||||
| Drug | Mode of Action | Comments | ||
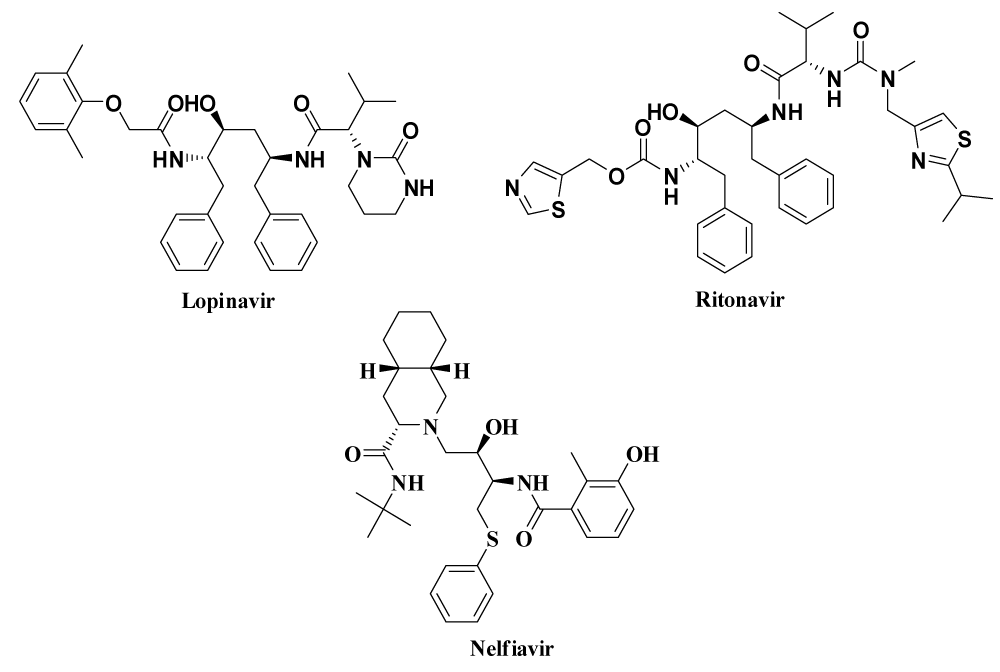
| Lopinavir & Ritonavir (ASC09F) [112-116] | Liponavir is a human immunodeficiency virus-1 (HIV-1) protease inhibitor and Ritonavir is a potent CYP3A4 inhibitor. Fixed-dose combination (LPV/r) is required to block the activity of protease enzyme and inhibiting viral replication. Herein ritonavir is boost up the activity of lopinavir. Lopinavir and ritonavir may bind to Mpro, a key enzyme for coronavirus replication. This may suppress coronavirus activity. | Abdominal pain, weakness, nausea, diarrhea, vomiting, headache and finally significant drug interaction. | ||
|
Inhibits the biological activity of protease namely, HIV-1 and HIV-2 proteases (aspartate protease) which can be restricted the replication of virus within the cell and also release the mature viral particles from the infected cell by splitting the viral protein molecules into smaller fragments. | No clinical data or in vitro analysis exist but based on the docking score and the 3D resemblance of binding mode, nelfinavir might be active against 2019-nCov. | ||
Figure 12: Drugs inhibiting virus non-structural proteins.
RNA-dependent RNA polymerase (RdRp) inhibitors: A preserved protein (Nsp12), RNA-dependent RNA polymerase (RdRp) in coronavirus, encode the vital enzyme to elaborate coronavirus replication/transcription complex. The RdRp domain of polymerase at the C-terminus has a conserved Ser-Asp-Asp motif. Nsp8 synthesizing up to six nucleotides in length can be used as a primer for Nsp12-RdRp RNA synthesis. Further, the Nsp7-Nsp8 complex improves the RdRps enzyme movement of Nsp12 along with binding to RNA. In the research of SARS-CoV inhibitors, Nsp12-RdRp has been used as a very important drug target. Inhibitory action and side effect of RdRp drugs are given in table 5 and their structures are shown in figure 13.
| Table 5; Drugs for inhibiting RdRp enzyme. | |||
| Sl. No. | Drug | Mode of Action | Comments |
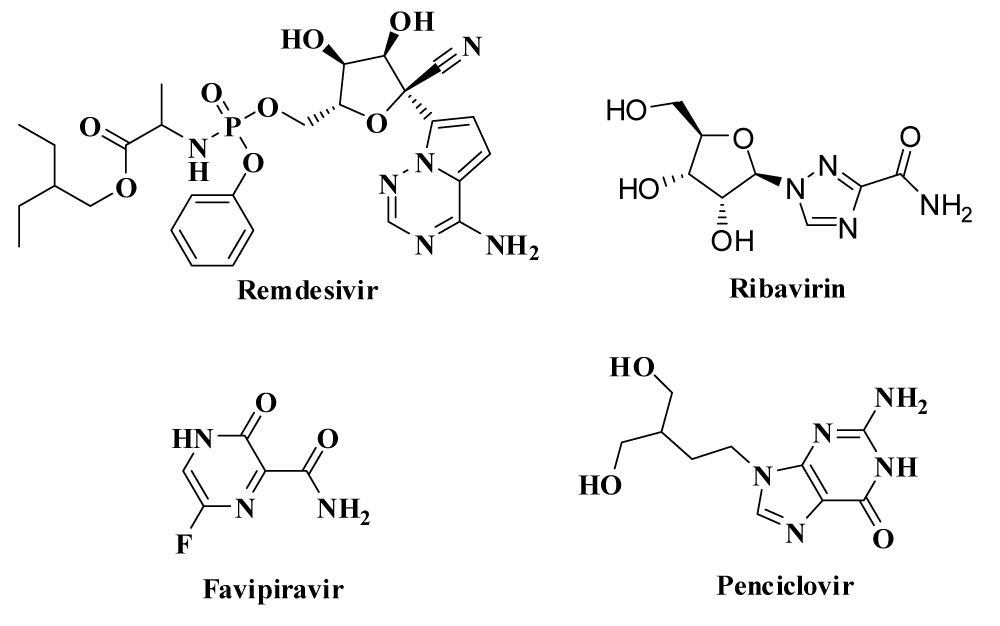
| 12.> | Remdesivir [96,118] (GS-5734)> | Remdesivir (GS-5734) is an investigational pro-drug developed by Gilead Sciences Inc. An adenosine nucleotide analogue, GS-441524 acts as an inhibitor of RNA-dependent RNA polymerases to terminate the chain formation of viral RNA, so that decrease the production of RNA. Since it is highly selective to viral polymerase, therefore must have low adverse effect in human cell. > | Affect to the liver by increasing the enzyme level which is reported in U.S. COVID-19 patients & also nausea, vomiting may occur. Potent in vitro activity against SARS-CoV-2 with an EC50 at 48 hours of 0.77 μM in Vero E6 cells. For treatment of COVID-19 one-time dose of 200mg intravenously Followed by 100mg per day for 10 days.> |
| 13.> | Ribavirin (tribavirin) [105,119-122]> | Ribavirin is a nucleoside inhibitor which is used to stop viral RNA synthesis and viral mRNA capping.> | Common side effects include feeling tired, headache, nausea, fever, muscle pains and an irritable mood. In trial period with the combination of pegylated interferon against COVID-19> |
| 14.> | Favipiravir ( Avigan) [96,119]> | Favipiravir is a pyrazine derivative which inhibits RNA elongation and chain termination by preventing the catalytic activity of viral RNA-dependent RNA polymerase enzyme.> | On 17 March 2020, Chinese officials suggested that Favipiravir seemed to be effective in treating COVID-19 in Wuhan and Shenzhen. > |
| 15.> | Penciclovir [105]> | Since Penciclovir is nucleoside analogue, exhibits low toxicity and good selectivity. It binds to the RdRp and terminates the replication process of viral RNA.> | Considered for clinical trial. > |
Figure 13: Drugs for inhibiting RdRp enzyme.
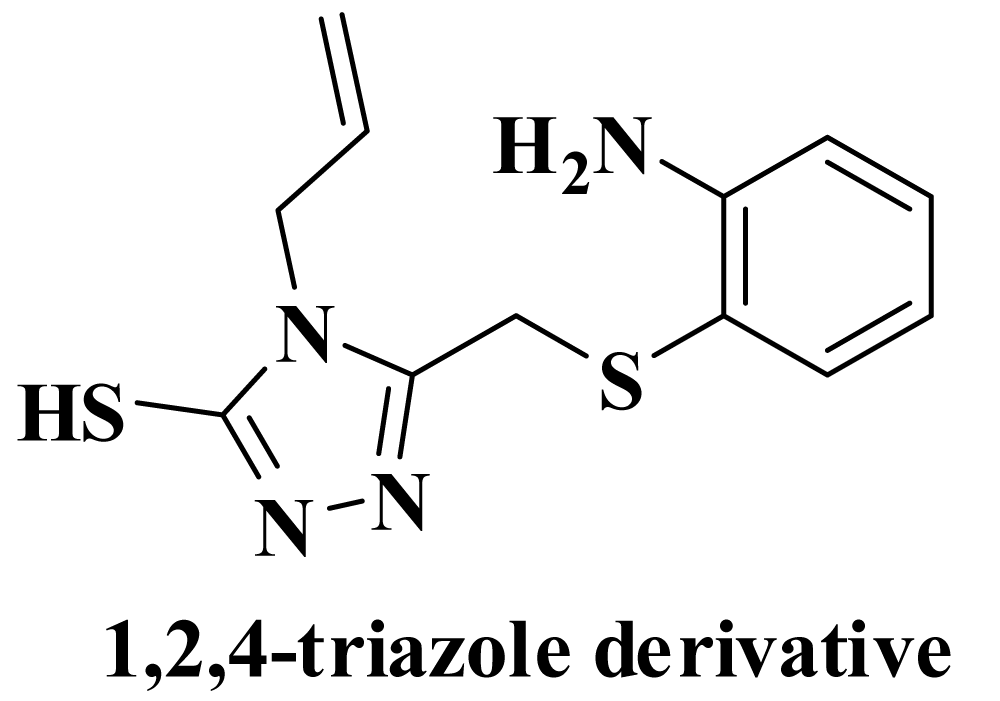
Drug targets inhibiting helicase: Helicase (Nsp13) plays a major role for virus replication in host cells by the process of N-terminal metal binding domain (MBD) and helicase domain (Hel) including downstream domain. Therefore, it can be chosen as anti-viral target for drug. Adedeji, et al. stated 1,2,4-triazole derivative which is shown in figure 14 behave as a helicase inhibitor. Some other drugs which can act as a helicase inhibitor are given in table 6.
Figure 14: Drug for inhibiting helicase.
| Table 6: Drugs for helicase inhibitor. | |||
| Sl. No. | Drug | Mode of Action | Comments |
| 16. | 1,2,4-triazole derivative [123-125] | Adedeji research group was reported 1,2,4-triazole derivative as the inhibitor of viral NTPase and helicase (NSP-13) of SARS- and MERS-Covs. | The antiviral activity of 87 inhibits MERS-CoV and SARS-CoV replication with EC50 values of 25 mM and 7 mM, respectively; and no significant cytotoxicity was observed up to the concentration of 500 mM. |
| 17. | Bananins and 5- hydroxyc-hromone derivatives[126] | Bananins and 5- hydroxychromone derivatives have selective inhibition activity against the helicase and ATPase | Preclinical |
Role of supporting drugs for COVID-19 treatment
Henceforth, there is no potential or effective the FDA approved drug for COVID-19. So it has become necessary to give supportive drugs along with the standard drugs. Table 7 lists some supportive medicines like immune-booster, anti-inflammatory, anti-biotic and active virus inhibitor.
| Table 7: Supporting drugs for COVID-19 treatments. | |||
| Sl. No. | Agent | Mechanism in Action | Comments |
| 18. | Tocilizu-mab[127] [127]128127126125125126 | Tocilizumab is a humanized monoclonal antibody used for the treatment of rheumatoid arthritis (RA). It inhibits the activity of interleukin 6 receptor (IL-6R) which is a pro-inflammatory cytokine that play a key role towards the pathogenesis of the virus and in immune response. | Upper respiratory tract infections, naso-pharyngitis, headache and high blood pressure. Standard dose is 4-8 mg/kg or 400mg dose IV once, with the option to repeat a dose in 12 hours (not to exceed a total dose of 800mg), recommended by National Health Commission of China. |
| 19. | Azithrom-ycin [128,129] | Macrolides are protein synthesis inhibitors and show their activity by prevents the growth of bacteria by interfering with their protein synthesis. It also inhibits the translation of mRNA through binding to the P site of the 50S subunit of the bacterial ribosome. Macrolides may also have immunomodulatory properties that mechanism involved inhibition of mucus hypersecretion, accelerating neutrophil apoptosis and reducing chemotaxis of neutrophils (PMNs) to the lungs by inhibiting cytokines (i.e. IL-8) [129-132]. | Diarrhea, stomach upset, nausea, abdominal pain and following risk factor-during the treatment of COVID-19 infected patients-cardiac arrhythmia and significant drug interaction. Pfizer has announced azithromycin as a supportive drug (with hydroxychloroquine) for COVID-19 infected patients in France. |
Drugs under investigation
In addition to the recommended and supportive drugs there are a number of drugs that are in clinical trials for the treatment of COVID-19. Table 8 illustrates some drugs which are under investigation or in trial period against COVID-19.
| Table 8: Drugs under investigation or in trial period against COVID-19. | |||
| Sl. No. | Agent | Mechanism of Action | Comments |
| 20. | Arbidol [133] (Umifenovir) | Inhibits membrane fusion to prevent the interaction between the virus (mainly RNA viruses) and target host cells. Therefore, the entry of virus into target host cells is inhibited and stimulated the immune response. | Not approved by the FDA but used as an Antiviral drug for influenza in Russia and China. Chinese clinical trials carried out for COVID-19 (200mg by mouth three times daily for no more than 10 days) claiming potent in vitro activity. |
| 21. | Olumiant [134] (Baricitinib) | Binds to Jenus Kinases type1 & 2(JAK 1/ 2) and leads to the inhibition of the JAK-signal transducers and activators of transcription (STAT) signaling pathway. Therefore, production of inflammatory cytokinaseis reduced and may prevent an inflammatory response. | Although no clinical data exist but artificial intelligence suggest Baricitinib for COVID-19 treatment. |
| 2. | Bevacizumab [135] (Avastin) | Inhibits the activity of vascular endothelial growth factor-A (VEGF-A) which is growth factor protein that stimulates angiogenesis in a variety of diseases and has a key role as potential therapeutic target in acute lung injury (ALI) and acute respiratory distress syndrome (ARDS). | It is a humanized anti-VEGF monoclonal IgG1 antibody. No clinical data exist but evaluated for clinical trial in China for COVID-19. |
| 23. | TZLS-501 [136] | IL-6 blocking pathway drug and IL-6 is considered as a key factor for chronic inflammation, associated with autoimmune diseases such as multiple myeloma, severe acute respiratory distress syndrome (ARDS). | China’s National Health Commission has suggested for the treatment of COVID-19 patients. |
| 24. | Sarilumab (Kevzara) [137,138] |
It is used for the treatment of rheumatoid arthritis (RA) by inhibiting the interleukin-6-receptor. | The Feinstein Institute of Northwell Health has recommended for the treatment of COVID-19. |
Vaccine Development for SARS-CoV-2
Development of vaccine typically required years of research and testing before reaching the clinic, but in 2020, enormous efforts of different scientists from different parts of the globe produced safe and effective coronavirus vaccines in record time. Researchers are currently testing 89 vaccines in clinical trials on humans, and 23 have reached the final stages of testing. At least 77 preclinical vaccines are under active investigation in animals. The first stage clinical trial for the vaccine was started in Wuhan on March 16, 2020 after approved authorisation. Spike protein of SARS-CoV-2 is the main part to involve the rapid development of specific vaccine [139]. The long-term goal of SARS-CoV-2 research is developing an effective vaccine to yield neutralizing antibodies. The National Institutes of Health in the US, and Baylor University in Waco, Texas, are working on a vaccine based on what they know about the coronavirus in general, using information from the SARS outbreak. In addition, the recent mapping of the SARS-CoV-2 spike protein may pave the way for more rapid development of a specific vaccine [139]. Furthermore, the production of RNA-based vaccines is more rapid and less expensive than traditional vaccines, which can be a major advantage in pandemic situations. Clinical trials for an mRNA-based SARS-CoV-2 vaccine are currently underway [140]. Recently, the vaccine of COVID-19 is based on the different part of the SARS-CoV-2 virus such as spike protein (S), envelope protein (E), nucleocapsid protein (N), N-terminal domain, membrane protein (M), whole cell of antigen, nucleic acid (DNA and RNA) [141]. Till now, no licensed vaccines are reported against SARS-CoV-2. Several research groups and institutes have tried to develop potential vaccine. It is reported that clinical trial (phase-I) will be conducted by the many research institutes based on different strategy like Pfizer, BioNTech (BNT162); Wuhan Institute of Biological Products; China National Pharmaceutical Group (Inactivated vaccine); Beijing Institute of Biological Products; China National Pharmaceutical Group (BBIBP-CorV); CanSino Biologics (Ad5-nCoV); Inovio Pharmaceuticals (INO-4800); Sinovac Research and Development Co., Ltd. (CoronaVac) [142]. University of Melbourne and Murdoch Children’s Research Institute; Radboud University Medical Center; Faustman Lab at Massachusetts General Hospital, The University of Oxford, the Jenner Institute are in Phase 2/3 trial period. A SARS-CoV-2 mRNA vaccine developed by Moderna, has in phase 2 trial period [142] (Table 9).
| Table 9: Development of vaccines from different parts of the world. | |||
| Development of vaccines | |||
| Developer | How does it Works | Phase | Status |
| Pfizer-BioNTech | mRNA | 2 and 3 | Approved in several countries. Emergency use in U.S., E.U., other countries. Clinical trials showed the vaccine has an efficacy of 95%. |
| Moderna | mRNA | 3 | Approved in Switzerland. Emergency use in U.S., E.U., other countries. |
| Gamaleya | Ad26, Ad5 | 3 | Early use in Russia. Emergency use in other countries. |
| Oxford-AstraZeneca | ChAdOx1 | 2 and 3 | Approved in Brazil. Stopped use in Denmark. Emergency use in U.K., E.U., other countries. |
| CanSino | Ad5 | 3 | Approved in China. Emergency use in other countries. |
| Johnson & Johnson | Ad26 | 3 | Emergency use in U.S., E.U., other countries. Paused in some states and countries. |
| Vector Institute | Protein | 3 | Early use in Russia. Approved in Turkmenistan. |
| Novavax | Protein | 3 | |
| Sinopharm | Inactivated | 3 | Approved in China, U.A.E., Bahrain. Emergency use in other countries. |
| Sinovac | Inactivated | 3 | Approved in China. Emergency use in other countries. |
| Sinopharm-Wuhan | Inactivated | 3 | Approved in China. Limited use in U.A.E. |
| Bharat Biotech | Inactivated | 3 | Emergency use in India, other countries. |
Since the very beginning Coronaviruses, have raised questions on therapeutic research and overall human health. Millions of people across the globe will have been infected with SARS-CoV-2 before its morphology is completely understood. Comparison of the reference genome of SARS-CoV-2, with the similar reference genomes of the coronaviruses discovered previously, will provide crucial information about the discovery of potential drugs and vaccine therapeutics. But these diagnostic steps (PCR, LAMP) are not quite effective and the actual treatment process with an exact drug is under clinical trials. With the passage of time, people have understood that a critical step like isolation is essential to prevent the spread of the COVID-19 pandemic. Even though the fatality rate of SARS-CoV-2 is less compared to SARS-CoVs and MERS-CoVs, the high transmission rate (R0) of SARS-CoV-2 makes it more perilous than the previous CoVs. Thus, an adequate number of detection kits must be available in time, to identify and isolate the infected patients from others, and to stop the nation-to-nation transmission rates. The outbreak of COVID-19 would not only affect human health but also have public repercussion, effecting negatively the cultural and socio-economic status of world, with an adverse effect specially on the tourism industry, logistic & retail markets, and in turn GDP of the countries. The crisis situation due to this sudden outbreak might not be overcome by the government and the public health authorities alone, unless people take immense care of themselves, by following the guidelines and policy decisions issued by the appropriate government and public health authorities to prevent the transmission of SARS-CoV-2. Obstructing the replication processes, like blocking of the interaction of spike glycoprotein of guest to the ACE2 or blocking the pathway for the synthesis of viral RNA by inhibiting the activity of RNA-dependent and RNA-polymerase enzyme of the host or binding of selected drug with the protease enzymes which are responsible for the synthesis of viral spikes, viral proteins in the host cell would become a potential molecular target to develop drugs therapeutics. Therefore, small molecular inhibitors and antibodies that can have the ability to achieve this are particularly interesting in the discovery of new drugs.
Although there are a few encouraging drugs mentioned in this study, the potential benefit of the proper drugs for management of treatment or prevention for COVID-19 infected, is yet to be defined, which results in a big therapeutic challenge. Therapeutic trials, protocols, warrant therapy and the timing of initiation, risk factors, progress rate, limitation, dose etc. on patients are still to be understood so that the medical practices continue to be updated time to time, from the data available or published by the government, or the FDA approved treatment protocol of COVID-19 to the community. Most importantly, there should be a strong requirement of quality-manpower for research and development of vaccine and potential drugs to fight against the fatal SARS-CoV-2. Therefore, all nations need to synergise research on this genome to achieve more robust results to fight against Coronaviruses.
The authors greatly acknowledge Dr. Mohabul Alam Mondal, Assistant Professor, Department of Chemistry, Jadavpur University for his helpful discussion and suggestions.
- Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020; 395: 470-473. PubMed: https://pubmed.ncbi.nlm.nih.gov/31986257/
- Lu H, Stratton CW, Tang YW. Outbreak of Pneumonia of Unknown Etiology in Wuhan China: the Mystery and the Miracle. J Med Virol. 2020; 92: 401-402. PubMed: https://pubmed.ncbi.nlm.nih.gov/31950516/
- Jiang F, Deng L, Zhang L, Cai Y, Cheung CW, et al. Review of the clinical characteristics of coronavirus disease 2019 (COVID-19). J Gen Intern Med. 2020; 35: 1545-1549.. PubMed: https://pubmed.ncbi.nlm.nih.gov/32133578/
- Tyrrell D, Bynoe M. Cultivation of viruses from a high proportion of patients with colds. Lancet.1966; 76-77. PubMed: https://pubmed.ncbi.nlm.nih.gov/4158999/
- McIntosh K, Dees JH, Becker WB, Kapikian AZ, Chanock RM. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc Nat Acad Sci U S A. 1967; 57: 933-940. PubMed: https://pubmed.ncbi.nlm.nih.gov/5231356/
- Bradburne A, Bynoe M, Tyrrell D. Effects of a" new" human respiratory virus in volunteers. Br Med J. 1967; 3: 767-769. PubMed: https://pubmed.ncbi.nlm.nih.gov/6043624/
- Stadler K, Masignani V, Eickmann M, Becker S, Abrignani S, et al. SARS—beginning to understand a new virus. Nat Rev Microbiol. 2003; 1: 209-218. PubMed: https://pubmed.ncbi.nlm.nih.gov/15035025/
- Peiris J, Lai S, Poon L, Guan Y, Yam L, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003; 361: 1319-1325. PubMed: https://pubmed.ncbi.nlm.nih.gov/12711465/
- Donnelly CA, Ghani AC, Leung GM, Hedley AJ, Fraser C, et al. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet. 2003; 361: 1761-1766. PubMed: https://pubmed.ncbi.nlm.nih.gov/12781533/
- Tahir M, Shah SIA, Zaman G, Khan T. A Dynamic Compartmental Mathematical Model Describing. The Transmissibility Of MERS-CoV Virus In Public. Punjab Univ J Math. 2019; 51 57-71.
- Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet. 2015; 386: 995-1007. PubMed: https://pubmed.ncbi.nlm.nih.gov/26049252/
- Lin L, Li TS. Interpretation of "Guidelines for the Diagnosis and Treatment of Novel Coronavirus (2019-nCoV) Infection by the National Health Commission (Trial Version 5)". Zhonghua Yi Xue Za Zhi. 2020; 100: E001. PubMed: https://pubmed.ncbi.nlm.nih.gov/32033513/
- WHO. WHO Director-General’s opening remarks at the media briefing on COVID-19–11.2020.
- Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020; 395: 470-473. PubMed: https://pubmed.ncbi.nlm.nih.gov/31986257/
- Ji W, Wang W, Zhao X, Zai J, Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol. 2020; 92: 433-440. PubMed: https://pubmed.ncbi.nlm.nih.gov/31967321/
- Priyanka, Choudhary OP, Singh I, Patra G. Aerosol transmission of SARS-CoV-2: The unresolved paradox. Travel Med Infect Dis. 2020. PubMed: https://pubmed.ncbi.nlm.nih.gov/32891726/
- Sahin AR, Erdogan A, Agaoglu PM, Dineri Y, Cakirci AY, et al. 2019 Novel Coronavirus (COVID-19) Outbreak: A Review of the Current Literature. EJMO. 2020; 4: 1-7.
- Dietz L, Horve PF, Coil D, Fretz M, Van Den Wymelenberg K. 2019 Novel Coronavirus (COVID-19) Outbreak: A Review of the Current Literature and Built Environment (BE) Considerations to Reduce Transmission. ASM J.2020; 5: e00245-20.
- Thompson R. Pandemic potential of 2019-nCoV. Lancet Infect Dis. 2020; 20: 280. PubMed: https://pubmed.ncbi.nlm.nih.gov/32043978/
- Leonardi D, Polidori C, Polidori P. The healthcare and pharmaceutical vulnerability emerging from the new Coronavirus outbreak. Eur J Hospital Pharm. 2020; 129-130. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7223278/
- Zaki AM, Van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New Engl J Med. 2012; 367: 1814-1820. PubMed: https://pubmed.ncbi.nlm.nih.gov/23075143/
- Chu H, Zhou J, Wong BHY, Li C, Cheng ZS, et al. Productive replication of Middle East respiratory syndrome coronavirus in monocyte-derived dendritic cells modulates innate immune response. Virology. 2014; 454: 197-205. PubMed: https://pubmed.ncbi.nlm.nih.gov/24725946/
- Li W, Shi Z, Yu M, Ren W, Smith C, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005; 310: 676-679. PubMed: https://pubmed.ncbi.nlm.nih.gov/16195424/
- Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013; 503: 535-538. PubMed: https://pubmed.ncbi.nlm.nih.gov/24172901/
- Luo H, Tang Ql, Shang YX, Liang SB, Yang M, et al. Can Chinese medicine be used for prevention of corona virus disease 2019 (COVID-19)? A review of historical classics, research evidence and current prevention programs. Chin J Integrat Med. 2020; 26: 243-250. PubMed: https://pubmed.ncbi.nlm.nih.gov/32065348/
- Lau JT, Leung P, Wong E, Fong C, Cheng K, et al. The use of an herbal formula by hospital care workers during the severe acute respiratory syndrome epidemic in Hong Kong to prevent severe acute respiratory syndrome transmission, relieve influenza-related symptoms, and improve quality of life: a prospective cohort study. J Altern Complement Med. 2005; 11: 49-55. PubMed: https://pubmed.ncbi.nlm.nih.gov/15750363/
- Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020; 38 1-9. PubMed: https://pubmed.ncbi.nlm.nih.gov/32105090/
- Mandal M, Chowdhury SK, Khan AA, Baildya N, Dutta T, et al. Inhibitory efficacy of RNA virus drugs against SARS-CoV-2 proteins: an extensive study. J Mole Struct. 2021; 1234: 130152. PubMed: https://pubmed.ncbi.nlm.nih.gov/33678903/
- Dutta T, Ghorai S, Khan AA, Baildya N, Ghosh NN. 2021 Screening of potential anti-HIV compounds from Achyranthes aspera extracts for SARS-CoV-2: An insight from molecular docking study. J Physics: Conference Series. IOP Publishing.
- Dutta T, Baildya N, Khan AA, Ghosh NN. Inhibitory effect of anti-HIV compounds extracted from Indian medicinal plants to retard the replication and transcription process of SARS-CoV-2: an insight from molecular docking and MD-simulation studies. Netw Model Anal Health Inform Bioinform. 2021; 10: 32. PubMed: https://pubmed.ncbi.nlm.nih.gov/33948424/
- Huang C, Wang Y, Li X, Ren L, Zhao J, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020; 395: 497-506. PubMed: https://pubmed.ncbi.nlm.nih.gov/31986264/
- Baildya N, Ghosh NN, Chattopadhyay AP. Inhibitory activity of hydroxychloroquine on COVID-19 main protease: An insight from MD-simulation studies. J Mole Struct. 2020; 1219: 128595. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7266611/
- Baildya N, Ghosh NN, Chattopadhyay AP. Inhibitory capacity of chloroquine against SARS-COV-2 by effective binding with angiotensin converting enzyme-2 receptor: An insight from molecular docking and MD-simulation studies. J Mole Struct. 2021; 129891. PubMed: https://pubmed.ncbi.nlm.nih.gov/33518803/
- Baildya N, Khan AA, Ghosh NN, Dutta T, Chattopadhyay AP. Screening of potential drug from Azadirachta Indica (Neem) extracts for SARS-CoV-2: An insight from molecular docking and MD-simulation studies. J Mole Struct. 2021; 1227 129390. PubMed: https://pubmed.ncbi.nlm.nih.gov/33041371/
- Khan AA, Baildya N, Dutta T, Ghosh NN. Inhibitory efficiency of potential drugs against SARS-CoV-2 by blocking human angiotensin converting enzyme-2: Virtual screening and molecular dynamics study. Microb Pathog. 2021; 104762. PubMed: https://pubmed.ncbi.nlm.nih.gov/33524563/
- WHO. Novel Coronavirus (2019-nCoV). Situation Report - 2. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200122-sitrep-2-2019-ncov.pdf
- Hui DSC, Zumla A. Severe Acute Respiratory Syndrome: Historical, Epidemiologic, and Clinical Features. Infect Dis Clin North Am. 2019; 33: 869-889. PubMed: https://pubmed.ncbi.nlm.nih.gov/31668196/
- Holshue ML, De Bolt C, Lindquist S, Lofy KH, Wiesman J, et al. First Case of 2019 Novel Coronavirus in the United States. New Engl J Med. 2020; 382: 929-936. PubMed: https://pubmed.ncbi.nlm.nih.gov/32004427/
- Wang D, Hu B, Hu C, Zhu F, Liu X, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020; 323: 1061-1069. PubMed: https://pubmed.ncbi.nlm.nih.gov/32031570/
- Torales J, Castaldelli-Maia JM, Ventriglio A. The outbreak of COVID-19 coronavirus and its impact on global mental health. Int J Soc Psychiatry. 2020; 66: 317-320. PubMed: https://pubmed.ncbi.nlm.nih.gov/32233719/
- Li Q, Guan X, Wu P, Wang X, Zhou L, et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. New Engl J Med. 2020; 382: 1199-1207. PubMed: https://pubmed.ncbi.nlm.nih.gov/31995857/
- Van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. New Engl J Med. 2020; 382: 1564-1567. PubMed: https://pubmed.ncbi.nlm.nih.gov/32182409/
- Van Zyl M, Fielding BC. The Coronavirus Nucleocapsid Is a Multifunctional Protein. Viruses. 2014; 6: 2991–3018. PubMed: https://pubmed.ncbi.nlm.nih.gov/25105276/
- Li F. Structure, function, and evolution of coronavirus spike proteins. Ann Rev Virol. 2016; 3: 237-261. PubMed: https://pubmed.ncbi.nlm.nih.gov/27578435/
- Guo Y, Korteweg C, McNutt MA, Gu J. Pathogenetic mechanisms of severe acute respiratory syndrome. Virus Res. 2008; 133: 4-12. PubMed: https://pubmed.ncbi.nlm.nih.gov/17825937/
- Luk HKH, Li X, Fung J, Lau SKP, Woo PCY. Molecular epidemiology, evolution and phylogeny of SARS coronavirus. Infect Genet Evol. 2019; 71: 21-30. PubMed: https://pubmed.ncbi.nlm.nih.gov/30844511/
- Coronavirinae in Viral Zone. 2019. https://viralzone.expasy.org/785
- Subissi L, Posthuma CC, Collet A, Zevenhoven-Dobbe JC, Gorbalenya AE, et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Nati Acad Sci. 2014; 111: E3900-E3909. PubMed: https://pubmed.ncbi.nlm.nih.gov/25197083/
- Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature.2003; 426: 450-454. PubMed: https://pubmed.ncbi.nlm.nih.gov/14647384/
- Te Velthuis AJ, van den Worm SH, Snijder EJ. The SARS-coronavirus nsp7+ nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012; 40: 1737-1747. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3287201/
- Stobart CC, Sexton NR, Munjal H, Lu X, Molland KL, et al. Chimeric exchange of coronavirus nsp5 proteases (3CLpro) identifies common and divergent regulatory determinants of protease activity. J Virol. 2013; 87: 12611-12618. PubMed: https://pubmed.ncbi.nlm.nih.gov/24027335/
- Wang H, Xue S, Yang H, Chen C. Recent progress in the discovery of inhibitors targeting coronavirus proteases. Virologica Sinica. 2016; 31: 24-30. PubMed: https://pubmed.ncbi.nlm.nih.gov/26920707/
- Egloff MP, Ferron F, Campanacci V, Longhi S, Rancurel C, et al. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc Nati Acad Sci.2004; 101: 3792-3796. PubMed: https://pubmed.ncbi.nlm.nih.gov/15007178/
- Hu T, Chen C, Li H, Dou Y, Zhou M, et al. Structural basis for dimerization and RNA binding of avian infectious bronchitis virus nsp9. Protein Sci. 2017; 26: 1037-1048. PubMed: https://pubmed.ncbi.nlm.nih.gov/28257598/
- Zhang M, Li X, Deng Z, Chen Z, Liu Y, et al. Structural biology of the arterivirus nsp11 endoribonucleases. J Virol. 2017; 91: e01309-13016. PubMed: https://pubmed.ncbi.nlm.nih.gov/27795409/
- Bouvet M, Lugari A, Posthuma CC, Zevenhoven JC, Bernard S, et al. Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. J Biolog Chem. 2014; 289: 25783-25796. PubMed: https://pubmed.ncbi.nlm.nih.gov/25074927/
- Narayanan K, Maeda A, Maeda J, Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J Virol. 2000; 74: 8127-8134. PubMed: https://pubmed.ncbi.nlm.nih.gov/10933723/
- de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016; 14: 523-534. PubMed: https://pubmed.ncbi.nlm.nih.gov/27344959/
- Nieto-Torres JL, DeDiego ML, Álvarez E, Jiménez-Guardeño JM, Regla-Nava JA, et al. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011; 415: 69-82. PubMed: https://pubmed.ncbi.nlm.nih.gov/21524776/
- Huang C, Wang Y, Li X, Ren L, Zhao J, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020; 395: 497-506. PubMed: https://pubmed.ncbi.nlm.nih.gov/31986264/
- Yang L, Tu L. Implications of gastrointestinal manifestations of COVID-19.Lancet Gastroenterol Hepatol. 2020; 5: 629-630. PubMed: https://pubmed.ncbi.nlm.nih.gov/32405602/
- Nguyen T, Duong Bang D, Wolff A. 2019 Novel Coronavirus Disease (COVID-19): Paving the Road for Rapid Detection and Point-of-Care Diagnostics. Micromachines. 2020; 11: 306. PubMed: https://pubmed.ncbi.nlm.nih.gov/32183357/
- Isere EE, Fatiregun AA, Ajayi IO. An overview of disease surveillance and notification system in Nigeria and the roles of clinicians in disease outbreak prevention and control. Niger Med J. 2015; 56: 161-168. PubMed: https://pubmed.ncbi.nlm.nih.gov/26229222/
- Jones G, Le Hello S, Jourdan-da Silva N, Vaillant V, De Valk H, et al. The French human Salmonella surveillance system: evaluation of timeliness of laboratory reporting and factors associated with delays, 2007 to 2011. Eurosurveillance. 2014; 19: 20664. PubMed: https://pubmed.ncbi.nlm.nih.gov/24434174/
- Nguyen T, Zoëga Andreasen S, Wolff A, Duong Bang D. From lab on a chip to point of care devices: The role of open source microcontrollers. Micromachines. 2018; 9: 403. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6187319/
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020; 25; 2000045. PubMed: https://pubmed.ncbi.nlm.nih.gov/31992387/
- Chu DK, Pan Y, Cheng SM, Hui KP, Krishnan P, et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020; 66: 549-555. PubMed: https://pubmed.ncbi.nlm.nih.gov/32031583/
- Bruning A, Aatola H, Toivola H, Ikonen N, Savolainen-Kopra C, et al. Rapid detection and monitoring of human coronavirus infections. New Micro New Infect. 2018; 24: 52-55. PubMed: https://pubmed.ncbi.nlm.nih.gov/29872531/
- Gaunt ER, Hardie A, Claas EC, Simmonds P, Templeton KE. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol. 2010; 48: 2940-2947. PubMed: https://pubmed.ncbi.nlm.nih.gov/20554810/
- Cho CH, Lee CK, Nam MH, Yoon SY, Lim CS, et al. Evaluation of the AdvanSure™ real-time RT-PCR compared with culture and Seeplex RV15 for simultaneous detection of respiratory viruses. Diagnostic Microbiol Infect Disease. 2014; 79: 14-18. PubMed: https://pubmed.ncbi.nlm.nih.gov/24582583/
- Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mole Cell Pro. 2002; 16: 223-229. PubMed: https://pubmed.ncbi.nlm.nih.gov/12144774/
- Galvez LC, Barbosa CFC, Koh RBL, Aquino VM. Loop-mediated isothermal amplification (LAMP) assays for the detection of abaca bunchy top virus and banana bunchy top virus in abaca. Crop Protection. 2020; 131: 105101.
- Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018; 360: 439-444. PubMed: https://pubmed.ncbi.nlm.nih.gov/29449508/
- Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020; 38: 870-874. PubMed: https://pubmed.ncbi.nlm.nih.gov/32300245/
- Drobysh M, Ramanaviciene A, Viter R, Ramanavicius A. Affinity Sensors for the Diagnosis of COVID-19. Micromachines. 2021; 12: 390. PubMed: https://pubmed.ncbi.nlm.nih.gov/33918184/
- Dronina J, Bubniene US, Ramanavicius A. The application of DNA polymerases and Cas9 as representative of DNA-modifying enzymes group in DNA sensor design. Biosens Bioelectron. 2020; 175: 112867. PubMed: https://pubmed.ncbi.nlm.nih.gov/33303323/
- Plikusiene I, Maciulis V, Ramanaviciene A, Balevicius Z, Buzavaite-Verteliene E, et al. Evaluation of kinetics and thermodynamics of interaction between immobilized SARS-CoV-2 nucleoprotein and specific antibodies by total internal reflection ellipsometry. J Colloid Interface Sci. 2021; 594: 195-203. PubMed: https://pubmed.ncbi.nlm.nih.gov/33761394/
- Mankar PSN. Review on recent emergence of novel corona virus 2019. World J Pharmaceut Res. 2020; 9: 2268-2273.
- https://www.aljazeera.com/news/2020/02/long-covid-19-outbreak-stay-protected-200212140216674.html?xif=.
- Komabayashi K, Seto J, Matoba Y, Aoki Y, Tanaka S, et al. Seasonality of human coronavirus OC43, NL63, HKU1, and 229E infection in Yamagata, Japan, 2010–2019. Japanese J Infect Dis. 2020; 525.
- https://www.nytimes.com/article/coronavirus-body-symptoms.html
- Geographical distribution of 2019- nCov cases. European Centre for Disease Prevention and Control data. An agency of the European Union. 2020.
- Li Q, Guan X, Wu P, Wang X, Zhou L, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. New Engl J Med. 2020; 382: 1199-1207.
- Novel Coronavirus. Prevention and Treatment. https://www.cdc.gov/coronavirus/2019-ncov/about/prevention-treatment.html.2019
- WHO. Report of the who-china joint mission on coronavirus disease 2019 (covid-19). Geneva: World Health Organization. 2020.
- Hui DS, EI A, Madani TA, Ntoumi F, Kock R, Dar O, et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020; 91: 264-266. PubMed: https://pubmed.ncbi.nlm.nih.gov/31953166/
- Arabi YM, Asiri AY, Assiri AM, Aziz Jokhdar HA, Alothman A, et al. Treatment of Middle East respiratory syndrome with a combination of lopinavir/ritonavir and interferon-β1b (MIRACLE trial): statistical analysis plan for a recursive two-stage group sequential randomized controlled trial. Trials. 2018; 19: 81. PubMed: https://pubmed.ncbi.nlm.nih.gov/29382391/
- Cockrell AS, Yount BL, Scobey T, Jensen K, Douglas M, et al. A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat Microbiol. 2016; 2 16226. PubMed: https://pubmed.ncbi.nlm.nih.gov/27892925/
- Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016; 531: 381-385. PubMed: https://pubmed.ncbi.nlm.nih.gov/26934220/
- WHO. Emergencies preparedness, response. Pneumonia of unknown origin – China. Disease outbreak news. https:// HYPERLINK"http://www.who.int/csr/don/12-january-2020-novelcoronavirus-china/en/"www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/ 2020
- Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003; 426: 450-454. PubMed: https://pubmed.ncbi.nlm.nih.gov/14647384/
- Yount B, Curtis KM, Fritz EA, Hensley LE, Jahrling PB, et al. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proceedings of the National Academy of Sciences.2003; 100 (22): 12995-13000. PubMed: https://pubmed.ncbi.nlm.nih.gov/14569023/
- Chen N, Zhou M, Dong X, Qu J, Gong F, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020; 395: 507-513. PubMed: https://pubmed.ncbi.nlm.nih.gov/32007143/
- Böttcher E, Freuer C, Steinmetzer T, Klenk HD, Garten W. MDCK cells that express proteases TMPRSS2 and HAT provide a cell system to propagate influenza viruses in the absence of trypsin and to study cleavage of HA and its inhibition. Vaccine. 2009; 27: 6324-6329. PubMed: https://pubmed.ncbi.nlm.nih.gov/19840668/
- Glowacka I, Bertram S, Müller MA, Allen P, Soilleux E, et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011; 85: 4122-4134. PubMed: https://pubmed.ncbi.nlm.nih.gov/21325420/
- Wu C, Liu Y, Yang Y, Zhang P, Zhong W, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B. 2020; 10: 766-788. PubMed: https://pubmed.ncbi.nlm.nih.gov/32292689/
- Gao J, Tian Z, Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Bioscience Trends. 2020; 14: 72-73. PubMed: https://pubmed.ncbi.nlm.nih.gov/32074550/
- Yao X, Ye F, Zhang M, Cui C, Huang B, et al. In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin Infect Dis. 2020; 71: 732-739. PubMed: https://pubmed.ncbi.nlm.nih.gov/32150618/
- German Researchers Identify Japanese Drug, Camostat Mesylate That Could Be Repurposed To Treat Covid-19. Coronavirus Drug Research. 2020. https://www.thailandmedical.news/news/coronavirus-drug-research-german-researchers-identify-japanese-drug,-camostat-mesylate-that-could-be-repurposed-to-treat-covid-19
- Ikeda S, Manabe M, Muramatsu T, Takarnori K, Ogawa H. Protease inhibitor therapy for recessive dystrophic epidermolysis bullosa: in vitro effect and clinical trial with camostat mesylate. J Am Acad Dermatol.1988; 18: 1246-1252. PubMed: https://pubmed.ncbi.nlm.nih.gov/3385039/
- Rossignol JF. Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus. J Infect Public Health. 2016; 9: 227-230. PubMed: https://pubmed.ncbi.nlm.nih.gov/27095301/
- Wang M, Cao R, Zhang L, Yang X, Liu J, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020; 30: 269-271. PubMed: https://pubmed.ncbi.nlm.nih.gov/32020029/
- Wu C, Liu Y, Yang Y, Zhang P, Zhong W, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B. 2020; 10: 766-788. PubMed: https://pubmed.ncbi.nlm.nih.gov/32292689/
- Ito K, Yotsuyanagi H, Sugiyama M, Yatsuhashi H, Karino Y, et al. Geographic distribution and characteristics of genotype A hepatitis B virus infection in acute and chronic hepatitis B patients in Japan. J Gastroenterol Hepatol. 2016; 31: 180-189.
- Asakura H, Ogawa H. Potential of Heparin and Nafamostat Combination Therapy for COVID-19. J Thrombosis Haemostasis. 2020; 18: 1521-1522. PubMed: https://pubmed.ncbi.nlm.nih.gov/32302456/
- Du L, Yang Y, Zhou Y, Lu L, Li F, et al. MERS-CoV spike protein: a key target for antivirals. Expert Opin Ther Targets.. 2017; 21: 131-143. PubMed: https://pubmed.ncbi.nlm.nih.gov/27936982/
- Lin SC, Ho CT, Chuo WH, Li S, Wang TT, et al. Effective inhibition of MERS-CoV infection by resveratrol. BMC Infect Dis. 2017; 17: 144. PubMed: https://pubmed.ncbi.nlm.nih.gov/28193191/
- Apaydın ÇB, Cesur N, Stevaert A, Naesens L, Cesur Z. Synthesis and anti‐coronavirus activity of a series of 1‐thia‐4‐azaspiro [4.5] decan‐3‐one derivatives. Archiv der Pharmazie. 2019; 352: 1800330. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7161747/
- Chen F, Chan KH, Jiang Y, Kao RY, Lu HT, et al. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J Clin Virol. 2004; 31: 69-75. PubMed: https://pubmed.ncbi.nlm.nih.gov/15288617/
- Yao TT, Qian JD, Zhu WY, Wang Y, Wang GQ. A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus-A possible reference for coronavirus disease-19 treatment option. J Med Virol. 2020; 92: 556-563. PubMed: https://pubmed.ncbi.nlm.nih.gov/32104907/
- Liu X, Wang XJ. Potential inhibitors for 2019-nCoV coronavirus M protease from clinically approved medicines. J Genet Genomics. 2020; 47: 119-121. PubMed: https://pubmed.ncbi.nlm.nih.gov/32173287/
- Chu CM, Cheng VC, Hung IF, Wong MM, Chan KH, et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004; 59: 252-256. PubMed: https://pubmed.ncbi.nlm.nih.gov/14985565/
- China. N H C N o t P s R o. The diagnosis and treatment guide of COVID-19 pneumonia caused by new coronavirus infection WHO. 2020; 7.
- Xu Z, Peng C, Shi Y, Zhu Z, Mu K, et al. Nelfinavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation. BioRxiv. 2020.
- Agostini ML, Andres EL, Sims AC, Graham RL, Sheahan TP, et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio. 2018; 9: e00221-00218. PubMed: https://pubmed.ncbi.nlm.nih.gov/29511076/
- Agostini ML, Andres EL, Sims AC, Graham RL, Sheahan TP, et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio.2018; 9: e00221-00218. PubMed: https://pubmed.ncbi.nlm.nih.gov/29511076/
- Al-Tawfiq JA, Momattin H, Dib J, Memish ZA. Ribavirin and interferon therapy in patients infected with the Middle East respiratory syndrome coronavirus: an observational study. Int J Infect Dis. 2014; 20: 42-46. PubMed: https://pubmed.ncbi.nlm.nih.gov/24406736/
- Arabi YM, Shalhoub S, Mandourah Y, Al-Hameed F, Al-Omari A, et al. Ribavirin and Interferon Therapy for Critically Ill Patients With Middle East Respiratory Syndrome: A Multicenter Observational Study. Clin Infect Dis. 2019; 70: 1837-1844. PubMed: https://pubmed.ncbi.nlm.nih.gov/31925415/
- Chan JF, Chan KH, Kao RY, To KK, Zheng BJ, et al. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J Infection. 2013; 67: 606-616. PubMed: https://pubmed.ncbi.nlm.nih.gov/24096239/
- Adedeji AO, Singh K, Kassim A, Coleman CM, Elliott R, et al. Evaluation of SSYA10-001 as a replication inhibitor of severe acute respiratory syndrome, mouse hepatitis, and Middle East respiratory syndrome coronaviruses. Antimicrob Agents Chemother. 2014; 58: 4894-4898. PubMed: https://pubmed.ncbi.nlm.nih.gov/24841268/
- Adedeji AO, Singh K, Calcaterra NE, DeDiego ML, Enjuanes L, et al. Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Antimicrobial agents and chemotherapy. 2012; 56: 4718-4728. PubMed: https://pubmed.ncbi.nlm.nih.gov/22733076/
- Zaher NH, Mostafa MI, Altaher AY. Design, synthesis and molecular docking of novel triazole derivatives as potential CoV helicase inhibitors. Acta Pharmaceutica. 2020; 70: 145-159. PubMed: https://pubmed.ncbi.nlm.nih.gov/31955138/
- Kim MK, Yu MS, Park HR, Kim KB, Lee C, et al. 2, 6-Bis-arylmethyloxy-5-hydroxychromones with antiviral activity against both hepatitis C virus (HCV) and SARS-associated coronavirus (SCV). Eur J Med Chem. 2011; 46: 5698-5704. PubMed: https://pubmed.ncbi.nlm.nih.gov/21925774/
- Megan Melody JN, Hastings J, Propst J, Smerina M, Mendez J, et al. Case report: use of lenzilumab and tocilizumab for the treatment of coronavirus disease 2019. Immunotherapy. 2020; 12: 1121–1126. PubMed: https://pubmed.ncbi.nlm.nih.gov/32546029/
- Clinicl Trials Arena. 2020. https://www.clinicaltrialsarena.com/news/pfizer-data-azithromycin-covid-19-trial/
- Gautret P, Lagier JC, Parola P, Hoang VT, Meddeb L, et al. .Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020; 105949. PubMed: https://pubmed.ncbi.nlm.nih.gov/32205204/
- Amsden GW. Anti-inflammatory effects of macrolides--an underappreciated benefit in the treatment of community-acquired respiratory tract infections and chronic inflammatory pulmonary conditions? J Antimicrob Chemother. 2005; 55: 10-21. PubMed: https://pubmed.ncbi.nlm.nih.gov/15590715/
- Beigelman A, Mikols CL, Gunsten SP, Cannon CL, Brody SL, et al. Azithromycin attenuates airway inflammation in a mouse model of viral bronchiolitis. Respir Res. 2010; 11: 90. PubMed: https://pubmed.ncbi.nlm.nih.gov/20591166/
- Zarogoulidis P, Papanas N, Kioumis I, Chatzaki E, Maltezos E, et al. Macrolides: from in vitro anti-inflammatory and immunomodulatory properties to clinical practice in respiratory diseases. Eur J Clin Pharmacol. 2012; 68: 479-503. PubMed: https://pubmed.ncbi.nlm.nih.gov/22105373/
- Boriskin Y, Leneva I, Pecheur EI, Polyak S. Arbidol: a broad-spectrum antiviral compound that blocks viral fusion. Current Med Chem. 2008; 15: 997-1005. PubMed: https://pubmed.ncbi.nlm.nih.gov/18393857/
- Richardson P, Griffin I, Tucker C, Smith D, Oechsle O, et al. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020; 395: e30-e31. PubMed: https://pubmed.ncbi.nlm.nih.gov/32032529/
- Bakri SJ, Snyder MR, Reid JM, Pulido JS, Singh RJ. Pharmacokinetics of intravitreal bevacizumab (Avastin). Ophthalmology. 2007; 114: 855-859. PubMed: https://pubmed.ncbi.nlm.nih.gov/17467524/
- https://www.tizianalifesciences.com/our-drugs/anti-il-6r/
- Iranbakhsh A. Potential applications of Plant Biotechnology against SARS-CoV-2. Iranian J Biol. 2021; 4: 89-98.
- Genovese MC, Fleischmann R, Kivitz AJ, Rell‐Bakalarska M, Martincova R, et al. Sarilumab plus methotrexate in patients with active rheumatoid arthritis and inadequate response to methotrexate: results of a phase III study. Arthritis Rheumatol. 2015; 67: 1424-1437. PubMed: https://pubmed.ncbi.nlm.nih.gov/25733246/
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020; 367: 1260-1263. PubMed: https://pubmed.ncbi.nlm.nih.gov/32075877/
- Paules CI, Marston HD, Fauci AS. Coronavirus infections—more than just the common cold. JAMA. 2020; 323: 707-708. PubMed: https://pubmed.ncbi.nlm.nih.gov/31971553/
- Zhang J, Zeng H, Gu J, Li H, Zheng L, Zou Q. Progress and prospects on vaccine development against SARS-CoV-2. Vaccines. 2020; 8: 153. PubMed: https://pubmed.ncbi.nlm.nih.gov/32235387/
- Craven J. COVID-19 vaccine tracker. Regulatory Affairs Professionals Society. 2021. https://www.raps.org/news-and-articles/news-articles/2020/3/covid-19-vaccine-tracker