More Information
Submitted: 13 July 2020 | Approved: 21 July 2020 | Published: 22 July 2020
How to cite this article: Raza ZH, Ihsan MA, Khan S, Zafar H, Rehman T. A Comprehensive review on genomic diversity and epidemiology of COVID-19. Int J Clin Virol. 2020; 4: 081-095.
DOI: 10.29328/journal.ijcv.1001021
Copyright License: © 2020 Raza ZH, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: COVID-19; Cardiovascular disease; Pregnancy; Infection; Transmission
A Comprehensive review on genomic diversity and epidemiology of COVID-19
Zeshan Haider Raza1*, Muhammad Ahmed Ihsan2, Sahrish Khan3, Haroon Zafar1 and Tayyaba Rehman1
1Institute of Biochemistry and Biotechnololgy, University of Veterinary and Animal Sciences, Lahore, Pakistan
2School of Agriculture and Veterinary Medicine, University of Padua, Padua, Italy
3Department of Biotechnology, Quaid-i-Azam University, Islamabad, Pakistan
*Address for Correspondence: Zeshan Haider Raza, Institute of Biochemistry and Biotechnololgy, University of Veterinary and Animal Sciences, Lahore, Pakistan, Email: [email protected]
A respiratory outbreak of COVID-19 started from Wuhan, China and on 30 January 2020, WHO declared this infection to be epidemic, implementing public health emergency worldwide. On 11th March 2020, observing its prevalence in the whole world and WHO declared as a pandemic. Many countries completely collapse in the grip of this pandemic, as there are no effective treatments available, the precaution is the sole remedy to minimize this infection. The emergence and pandemic of SARS-CoV-2 (since the SARS-CoV in 2002 and MERS-CoV in 2012] manifest the third time outline of highly contagious and pathogenic infection with infect-ability to spread globally in the twentieth-first century. The SARS CoV-2 genome is highly identical to bat coronavirus which is considered to be the perfect natural host. This coronavirus even utilizes the same ACE2 receptor as SARS-CoV and mainly spread the infection to the respiratory tract, which evidently showed that transmission of this virus through interactions and exposures. The death toll of these infected patients is increasing day by day especially when they have prehistory fatal diseases like cardiovascular, diabetics, and respiratory diseases. In this review, we summarized and explained the research progressed and available data on epidemiology, COVID-19 phylogenetic relation and its impact of different fatal disease and their relation and discuss the precautionary methods to combat this pandemic. Moreover, the pieces of evidence of spreading the virus through pets and prevention of being spreading by copper metal endorsement.
At the end of 2019, almost 27 cases having the same symptoms pneumonia identified in Wuhan city in China, arose from the Human Seafood Wholesale market [1-3]. Only after the investigation by Chinese officials, it was confirmed that the cluster and symptomology have been associated with the Novel Coronavirus 19, and WHO gave the official name as COVID-19 [4]. After the phylogeny and homology analysis of COVID-19, it is confirmed that the COVID-19 is very closely associated with Bat origin SARS-CoV (88% identity) [5-7]. On 11, March 2020, WHO declared the COVID-19 infection as pandemic and up-to-date of 15 April 2020, 2.034 million infected patients all over the world spreading in over 180 countries with a total 129,951 (6.3%) deaths and total 496,562 (24.4%) recovered [8].
To date most of the patients who are infected with this virus develop mild symptoms such as sore, dry cough, fever, and throat [9]. Most of the cases resolved easily but some cases due to multiple complications, develop the fatal reactions including septic shock, severe pneumonia, ARDS (Acute Respiratory Distress Syndrome), organ failure and pulmonary edema [6,10-12]. Most of these patients either have some history with diabetes, endocrine, respiratory and cardiovascular issues, or have a weak immune system and these patients require intensive care support [13-17]. So, far there is no specific, proven or effective pharmaceutical treatment available [12,17-21]. The aim of this review is to gather the latest data available regarding COVID-19 infection and its relation to many disorders which cause serious condition patients, so further research will be performed [11,22].
The information for the article was collected by two sources, offline and online. Both of the sources proved useful, practical, and beneficial for collecting data. The first type of sources (offline sources) includes the diversity of relevant books from the library, the proceeding of seminars and the abstract booklets of the conferences. The second type of source is an online source which is mostly contain digital data from the internet. This type of data includes the original research articles, scientific reports, published review articles, official websites of different organization and newsletters in well-known reputed journals. A total of 119 Articles (including review and original) were retrieved from the google scholar, sciences direct and other scientific data search engines. A good variety of reputed and trustworthy publishers was also used to collect the data. A large proportion was collected from Elsevier and Willy’s online library. Different keywords like SARS CoV-2, Pregnancy effect of COVID-19, Cardiovascular disorder and COVID-19, Phylogenetics relations, Transmission procedure, epidemiology and other keywords were used so that only relevant data can be collected with little effort and to obtain time efficiency. Some of the reputed impacts factored journals like “The Lancet”, “International Journal of Infectious Diseases”, “Jama” and “Nat Med” were explored to gather relevant valuable information for this article.
Some valuable research articles haven’t open access to the readers. Such articles were requested for full-length free PDF form by the UVAS (the University of Veterinary and Animal Sciences, Lahore) E-library to the corresponding journals and publishers. This sponsor aided the writer a lot to obtain a well-shaped review article. After collecting and arranging all the data for the review paper, it was further proofread by all of the co-authors and necessary changes were made. The information collected from online sources were rephrased to avoid plagiarism concerns.
Novel coronavirus structure and genomic analysis
Coronaviridae is the family of viruses that mostly found within fish, birds as well as mammals [10,23]. The molecular characterization of the coronavirus was made in 1960. In Himalayan palm civets, the firstly discovered [24]. Respiratory infection is the major disease caused by this virus especially in children and older-aged (above 60 years) [21]. The virus contains 29 unique additional nucleotides that are not found in humans. Later, the same type of virus was being isolated from horseshoe bats [25,26]. If the bat-SARS-CoV genome is being added by 29 base pairs extra then this will make a similar virus structure that is responsible for the COVID-19 in humans [26]. WHO classified COVID-19 as a 2B group further divides as β CoV [16]. Ten samples of COVID-19 from different patients were collected and sequenced which showed that it is 99.98% similarity between them [6,27]. The very first discovered coronavirus was the Infectious bronchitis virus (IBV) and it was isolated from a chick embryo in 1937 [28]. Later, in the 1960s, the two more types of coronaviruses were discover named human coronavirus 229E (HCoV-229E) and human coronavirus OC43 (HCoV-OC43] [29,30]. There are basically three groups of coronaviruses named groups 1, 2 and 3 respectively. Groups 1 and 2 contain the 9 mammalian coronaviruses and group 3 includes the avian coronavirus [25,26]. Up to 2008, during the SARS epidemic, there are 16 more coronaviruses were sequenced as a whole genome. This includes the two worldwide spreading human coronaviruses, which are (HCoV-NL63] and HCoV-HKU1 [18, 31-38]. The 10 more coronaviruses in mammals which are bat-CoV 512/2005bat-CoV HKU8, bat-CoV HKU9, bat-CoV 1A, equine coronavirus, bat-CoV HKU5 and four coronaviruses in birds [15,16,35-40]. furthermore, this is divided into two more subgroups in group 2 coronavirus that is 2c and 2d and similarly two subgroups in group 3 coronavirus which are 3b and 3c [25,32,35,41,42].
SARS CoV-2
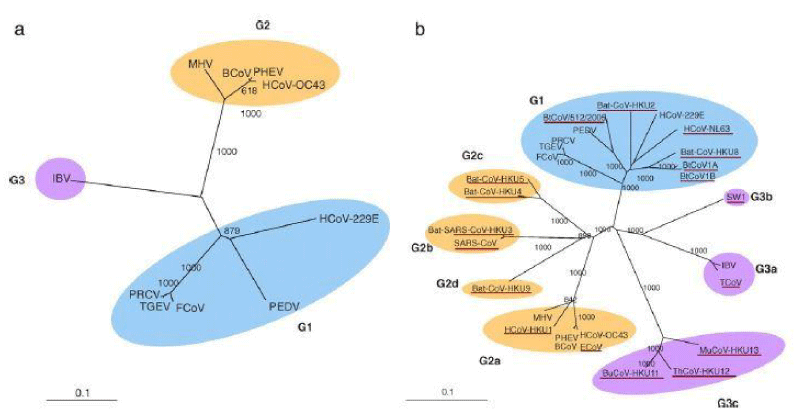
The phylogenetic tree of RNA-dependent RNA polymerases (Pol) obtaining from 10 different coronaviruses using whole sequenced genome that were available before SARS (panel A). These complete genomes were sequenced until the end of 2008 (panel B). The tree was constructed by using the option of the Neighbor-joining method using Kimura’s two-parameter correction command. The bootstrap value was being set at 1000 trees as shown in figure 1.
Figure 1: Phylogenetic analysis of RNA-dependent RNA polymerases (Pol) of the 10 coronaviruses with complete genome sequences available before SARS (panel A), and that of all coronaviruses with complete genome sequences available by the end of 2008 (panel B) (17).
By interpreting the phylogenetic tree, it was revealed that the SARS COV-2 is from to genus beta-coronavirus that includes SARS CoV which can infect human, bat, and wild animals. SARS COV-2 is classified as the 7th member of the coronavirus family. It particularly has the tendency to infect humans and it included in the orthocoronavirinae subfamily. SARS COV2 (responsible for pandemic COVID-19] forms a clade lies the subgenus sarbecovirus [10,23]. The prevalence of greater similarity of the ACE2 receptor showed the evidence that this virus is from the bat origin. It means the animal species can be the host of this virus, most probably, for COVID-19 infection [43]. The ORF (open reading frame) of this virus is at gene number 8 that is also heavy evidence of its origin from the bat. Another indicator that SARS COV-2 has the bat origin is the resemblance of an amino acid sequence of the receptor-binding domain with SARS-CoV [13,44].
Phylogenetic analyses of coronavirus and comparison with other strains
SARS COV2 has a great value of resemblance to the Bat-CoV RaTG13 which was isolated from the bats in the Yunnan Province of China. RaTG13 is the closest to the sequence of SARS COV2, almost throughout the genome. It has almost 96.35 % homology (p -distance: 0.0369). As it has a concern with 5’-part of the complete genome, first 10,901 nucleotides within alignment, which are related to the 11,498 nucleotides of the prototype strain (NC_045512]. The phylogeny revealed that the 3’-part of the genome at the position of 22,831-27,933 (24,341-30,696 nucleotides in the NC_045512), 2019-nCoV and RaTG13 lies in a common cluster with coronavirus sequences related to Bat SARS. As the middle part of the genome is a concern, the region spanning the 3’-end of the ORF1b, ORF1a, and somehow the half of the spike regions [10,901-22,830 nucleotides in the alignment or 11,499-24,340 of the NC_045512], 2019-nCoV and RaTG13 grouped in a separate distant lineage lies in the sarbecovirus clade.
Unlikely, in the middle region, there a no similarities found between the 2019-nCoV and RaTG13 with relation to Bat-SARS-like coronavirus genome sequences. Moreover, the BLAST search of the genome of 2019-nCoV was carried out from the middle fragments, which elucidated that the middle portion is dissimilar with respect to previously sequenced coronaviruses. The sequence comparison revealed that there is 88% – 92% similarity of nucleotide between SARS-CoV-related coronaviruses isolated from horseshoe bats and human/civet SARS-CoV. The most important part of the study revealed that the spike (S) proteins of a bat as well as of human viruses were found that there is 78% – 80% homology of the nucleotide sequence [25,26,41,45].
An independent novel group of human CoVs (HCoVs) was classified besides the existing two groups in 2004. Molecular characterization, molecular epidemiology, clinical characters, and whole-genome sequence of unique group 2 were reported in 2005 [13,25,26,31,44]. Further studies on coronavirus led the scientist to find 9 more coronaviruses consist of two new subgroups of coronaviruses named groups 2c and 2d [16,37,38,46]. Moreover, it was revealed from a study that SARS-COV-2 has 80% homology with already existing human coronavirus [6,47]. While doing basic research, it was revealed by the evolutionary tree that SARS-CoV-2 has a very close relation to the other groups of SARS-coronaviruses [6]. But new researches confirmed that there is a significant difference between SARS-CoV and SARS-CoV-2.
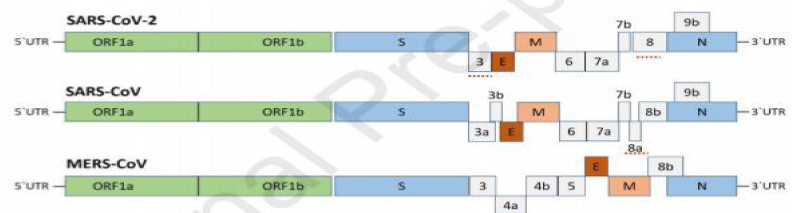
One of the major differences is the lack of 8a protein as well as variability in the amino acid count in 8b as well as in 3c protein in SARS-CoV-2. The studies revealed that the phenomena of homologous recombination change the spike glycoprotein in the coronavirus strain of Wuhan city. Bat SARS-CoV and an unknown Beta-CoV protein combined to form glycoprotein of SARS-CoV-2 60). In a study based on fluorescent technique, it has been confirmed that the SARS-CoV-2 and the SARS-CoV have a common receptor ACE2 (angiotensin-converting enzyme 2) receptor of a cell and shared the mode of entrance in a host [48,49]. The binding affinity for ACE2 can be increased by just single nucleotide polymorphism in SARS-CoV-2’s Spike protein.
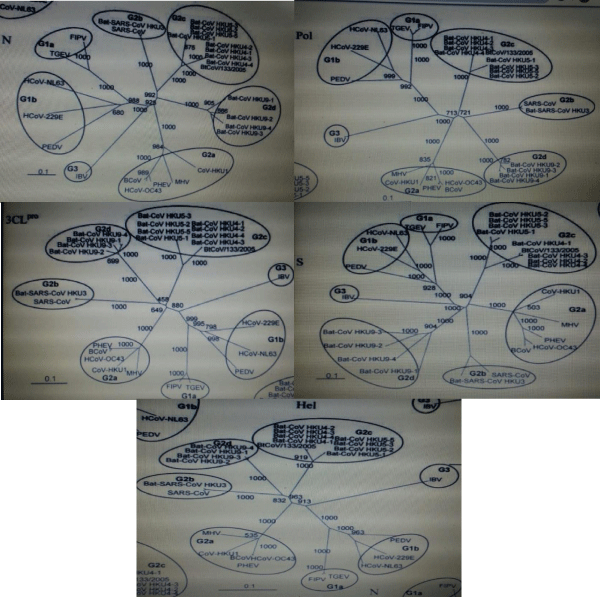
In a study phylogenetic analysis helicase as seen in figure 2, S protein, 3CLpro, Pol and N of bat-CoV HKU4, bat-CoV HKU9, bat-CoV HKU5, and other coronaviruses was carried out by maximum likelihood method with respect to pairwise amino acid identities. All genes including bat-CoV HKU5, bat-CoV, HKU4, and bat-CoV HKU9 have a high value of similarities as compared to group 2 coronaviruses. In the next step, it was compared with group 1 and group 3 coronaviruses repressively. The constructed trees showed that all strains of bat-CoV HKU5, bat-CoV HKU4, and recently discovered strains were made the same clade.
Figure 2: Phylogenetic analysis of Chymotrypsin-like-protease (3CLpro), RNA-dependent RNA polymerase (Pol), Helicase (Hel), Spike (S), and nucleocapsid (N) of bat-CoV HKU4, bat-CoV HKU5, and bat-CoV HKU9 (12).
In all trees, the bootstrap value was set as 1000 which resulted in the formation of the novel subgroup. This subgroup was containing the four-strain of bat-CoV HKU4 and all were found clustered at the same clade with the recently known strain explained in figure 3 (BtCoV/133/05) [50,51]. And the strains of bat-CoV HKU5 made a different clade by the formation of two additional sub-lineages. Moreover, the five trees of all strains of batCoV HKU9 formed a single common cluster and in all trees, the bootstrap values were taken as 1,000. This resulted in the formation of another new subgroup. The group 2b coronaviruses and the strains of the bat-CoV HKU9 subgroup were found very close to each other as compared to all others. There were two distinct groups were thought to be in existence as group 2c and group 2d.
Figure 3: The bootstrap value was set as 1000 which resulted in the formation of the novel subgroup. This subgroup was containing the four-strain of bat-CoV HKU4 and all were found clustered at the same clade with the recently known strain [17].
The phylogenetic comparison was carried out between helicase (Hel), spike (S), RNA polymerase (Pol), chymotrypsin-like protease (3CLpro), bat-HKU9, bat-CoV HKU4, bat-CoV HKUS nucleocapsid (N). By using neighbor-joining methods with 1000 bootstraps value and Juke-cantor correction, the tree was contrasted. After evaluating results, the 3CL, helicase, Pol, S, and N came at amino position 327, 609, 949, 1661, and 582. The scale bar is showing the estimated number of changes per 10 amino acids [17,40].
The 22 CoV HKU1 strains of coronaviruses contained the size ranging from 29,295 to 30,097 nucleotides. And the GC contents percentage is almost 32. It is confirmed from the studies that all 22 CoV HKU1 strains have the same genome structuring. Phylogenetic analysis was carried out between sequences of putative polypeptide genes and some proteins such as conserved nsp3 portions, nsp2, nsp1 [A1pp, PL1pro, HD, and PL2pro], M, N E, nsp12-nsp16, nsp4-nsp10, S and ORF4 HE of CoV HKU1 strains. The three clusters are being formed in 18/24 trees by overall 22 CoV strains with having named genotype C (6 strains), Genotype B (3 strains), genotype A (13 strains). The five trees made by using nsp1, nsp2, PL1pro, PL2pro, and HD, which contained very little difference and the nsp10 tree, in which two genotypes A strains, N1 and N3, were made a clustered with genotype C strains. The sequences of putative transcription regulatory motif which are 5-A AUCUAAAC-3 (as in mouse hepatitis virus (MHV) and bovine coronavirus) [15,32,52] or alternatively, 5-UAAAUCUAAA C-3, were seemed at the 3 ends of the leader sequence and preceded each translated Open Reading Frame except ORF5 which are found in the genome of the CoV HKU1 genotype A strain which can be instituted by all CoV genomes. The IRES (Internal ribosomal entry site) [26,36] have the same ORF envelope protein in genotype B strains in all 3 genomes while in genotype A have AUUUAUUGUUUGG instead genotype C strain have the UUUUAUCGCUUGG.
However, both sequences have somehow similarity to the IRES element, UUUUAUUCUUUUU, in MHV. The repeating sequences of 30 base pairs acidic tandem repeat (ATR) in the N terminus of nsp3 were varied in 22 genome sequences at the upstream of PL1pro. The encodes NDDEDVVTGD, as well as different numbers of imperfect repeats, were found the 30-base pair tandem repeats in all of 22 genomes which were studies. The imperfect repeats showed a median of 2 ranging from 1 to 4. On the other hand, the perfect repeats bear the median number of 11.5 ranging from the 2 to 17 in the copes of 30 base pairs. The 10 CoV HKU1 strains bearing not complete repeats having imperfect repeats of 1.4 and 4.4, were from the genotype A [10,15,36].
In a study, a total of 53 SARS-CoV-2 complete sequences documented up to February 3, 2020, were studied. There was a clear indication of the relation of sample collecting time and the mutation pattern of SAR-COV-2. It was analyzed by phylogenetic analysis that the virus that was found in Wuhan (China) was introduced in December 2019. In this analysis, Bayesian and maximum likelihood phylogenetic methods were used to construct the tree and found that the epidemic doubling time of virus was almost 7 days. The exact epidemic size of SARS-Cov-19 is not still known with available data. The studies show secondary infection has heterogeneity in the values of secondary infections and the data shows the high level of dispersion in the case of COVID-19 [53]. The previously identified coronaviruses (SARS-like bat CoV) in humans and SARS-Cov-2 has almost 80% similarities in their genetic makeup [32]. There are four main proteins that form the structure of coronavirus that includes S protein forms petal-shaped surface projection; other proteins encode envelope (E protein); a membrane glycoprotein and a nucleocapsid protein that forms a complex with RNA. The genes are known as Spike (S), envelope (E), membrane (M), and nucleocapsid (N) genes. The largest gene in SARS-CoV-2 is orf1ab. This gene encodes pp1ab protein as well as 15 nsps. The orf1a gene that encodes pp1a protein and 10 nsps [38,54-56]. The evolutionary tree the novel coronavirus has greater resemblance with the group of SARS-coronaviruses [6,48].
Phylogenetic tree of novel coronaviruses
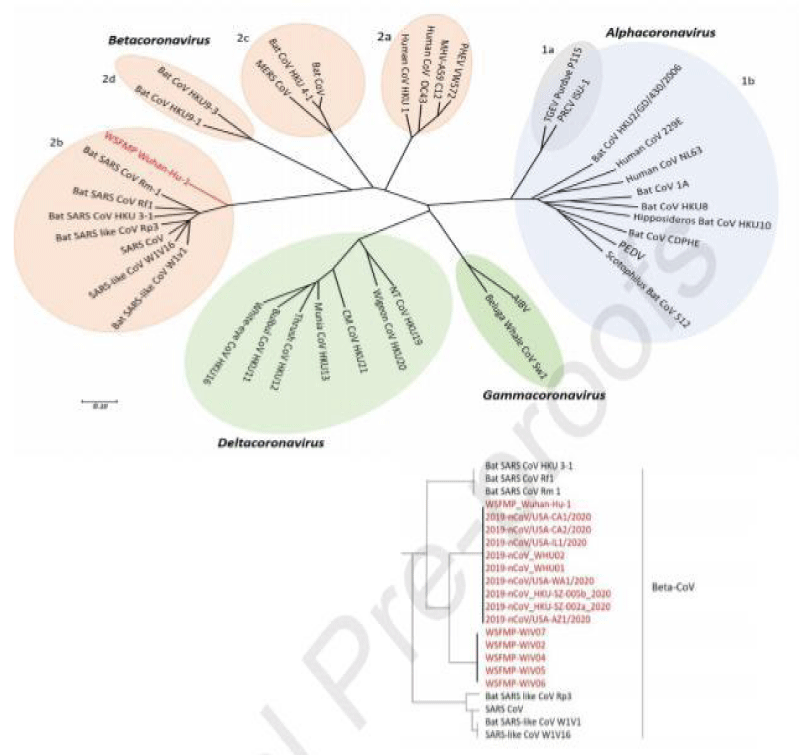
Phylogenetic analysis of coronaviruses was carried out in which the novel SARS-CoV-2 and WSFMP Wuhan-Hu-1 are taken as a reference showed in red color as shown in figure 4. The tree is showing the relation of Wuhan-Hu-1 (indicated as red) and other selected coronaviruses. The whole-genome sequences were used in this tree. The viruses were categorized into four types (prototype shown): Alpha-coronavirus (sky blue), Beta-coronavirus (pink), Gamma-coronavirus (green), and Delta-coronavirus (light blue). Subgroup clusters can be seen as 1a and 1b for the Alpha-coronavirus and 2a, 2b and 2c, and 2d for the Beta-coronavirus. The tree was reconstructed from earlier published trees of Coronaviruses with maximum likelihood method using MEGA 7.2 software) [49,57,58]. SARS- related coronavirus (SARSr- CoV); the Middle East respiratory syndrome coronavirus (MERS- CoV); Severe acute respiratory syndrome coronavirus (SARS- CoV); Wuhan seafood market pneumonia (Wuhan-Hu-1]; porcine enteric diarrhea virus (PEDV). Bat CoV RaTG13 Showed high sequence identity to SARS-CoV-2 [59].
Figure 4: Phylogenetic tree of coronaviruses (content in red is the latest addition of newly emerged SARS-CoV-2 and WSFMP Wuhan-Hu-1 is used as a reference in the tree) [12].
Pets with COVID-19
A pet dog in Hong Kong was tested positive with COVID-19 on February 26th, 2020. And one day earlier on 25th, the owner of the dog was also reported positive for COVID-19. In both, the genetic makeup of the virus was found almost similar. This became a mystery that whether the disease was transferred from owner to dog or dog to owner. After hospitalization both, dog and owner were tested negative after the required quarantine period. But just two days after the discharge from the hospital, the dog was found dead. This dilemma made the scientist curious about the cause of death. Only one answer was given by some animal welfare experts that the reason for death was the stress during quarantine and maybe being separated from the owner [60]. But this is still a debatable topic.
Furthermore, according to the World Health Organization, Centre for Disease Control and Prevention and World Organization for Animal Health has reported on the 2nd of April, 2020 that there is negligible evince found for the transmission of COVID-19 from pets to humans. But still, all organizations advised staying separate from the pets until the finalization of research. The SARS COV-2 is the animal origin virus so animals are thought to be an efficient host of the virus [61].
In a recent report published on April 6, 2020, the United States Department of Agriculture (USDA) National Veterinary Services Laboratories has confirmed the first positive case of COVID-19 in the tiger. The tiger was kept in the zoo in New York. Many other lions and large cats were showing the same symptoms but still not tested positive. Authorities confirmed that the virus was transferred from a worker (having COVID-19 positive) to the tiger. But still, research is ongoing to solve this situation [62].
It is reported that SARS-CoV-2 replicates poorly in dogs, pigs, chickens, and ducks, but efficiently in ferrets and cats. It was found in studies that the virus transmits in cats via respiratory droplets [60].
The Mustela furo and Felis domesticus are more susceptible to SARS COV infections. And these types of viruses can be transmitted from one animal to another or to the humans who are living together in homes (pet owners) [63].
On the other hand, a study was conducted to elucidate whether the Middle East respiratory syndrome (MES) can be transmitted from camel to humans or not. The serum samples were analyzed. A total of 191 human samples were collected with different levels of time exposure with the infected herd. This study concluded that transmission of the virus is very rare from camel to humans [64].
Transmission and epidemiology
This SARS-CoV-2 is a β-coronavirus, non-segmented positive RNA virus, enveloped, belongs to Orthocoronavirinae subfamily. The division of CoV is done in four groups, including α−/ β−/γ−/δ-CoV. α- and β-CoV capable of causing infection to mammals, while the δ- and γ-CoV cause infection in birds [65]. There is a total of six CoVs have identified and among them two SARS-CoV and MERS-CoV conduct a severe and serious fatal respiratory infection [66,67]. After genome sequencing, the SARS-CoV-2 found to 96.2% identical and similar to bat CoV RaTG13, while for SARS-CoV it was 79.5% [68,69]. Getting the details of the virus genome, the results clearly show that the suspected natural host of this virus origin is Bat, but the transmission of a human through intermediate host which is still unknown. After researching, the phenomena are clear that SARS-CoV-2 use angiotensin-converting enzyme 2(ACE2), the receptor is the same for an existing outbreak from the same Coronavirus to cause infection to human [65-67].
On 12 December 2019, the epidemic outbreak arose having acute respiratory symptomology from Wuhan, China in a seafood market [70,71]. Multiple studies had been performed to check and trace the potential source and found that the potential, thou no evidence was found but the fact the bat host variety of CoVs including MERS-CoV and SERS-CoV [72]. After the genome sequence analysis and 96.3% match give us some insight that human SARS-CoV-2 and bat CoV might share the same ancestry. Besides, the protein sequence analysis gives more details possibility about the intermediate host which might be pangolin, turtles, and snacks [65,73].
Besides the animal to humans, there is human to the human transmission which makes this outbreak from epidemic to pandemic. As the virus cause acute respiratory infection and multiply in the respiratory tract, so during breathing it transfer the infection to a nearby person. SARS-CoV-2 is an airborne virus, transmit between family members, friends, colleagues, who make contact with incubation carrier or patients [67]. During this outbreak, 72.3% of patients spread the virus from Wuhan to other state and 31.3% of patients traveled and spread this virus to the whole Wuhan region and outside to it. The transmission of COVID-19 from patients to healthcare workers with 3.8% as data stated by NHC china. Till April 15, 2020 total of 82,295 confirmed cases were reported with a mortality rate of 4% and total recovered patients of 77,816 [94%). The transmission of MERS-CoV and SARS-CoV was mainly nosocomial, while for SARS-CoV-2 transmission route is direct contact, through respiratory droplets from sneezing and coughing [67,72].
Per epidemiology, the numbers of cases which is provided by WHO Health Emergency Dashboard [15 April 2020, 10:00 CET), the overall number rose over 2.034 million infected patient all over the world spreading in over 180 countries with total 129,951 (6.3%) deaths and total 496,562 (24.4%) recovered. Among these numbers, the United States of America has the highest number of cases which is 578,268 with 23,476 deaths (4%), Spain with [177,633], Italy [165,155] and France [143,303] and every day, the number of the infection rose in the whole world. However, with so modern technology, the transmission and source of SARS-CoV-2 remain elusive [8,20].
Symptoms of COVID-19
The disease caused by the novel coronavirus (SARS-Cov-19] sometimes shows some symptoms and sometimes there is no symptom found in active cases. Almost 1% of patients showed no proper symptoms of this disease [7]. Still, there are some symptoms that are noted from the diagnosis of patients. The clinical symptoms are mostly characterized by shortness and difficulty inbreeding. Furthermore, there is an infection in an upper and lower respiratory tract is noted with complete organ failure. Dyspnea, malaise, high fever, and dry cough are also seen in the patients of COVID-19 [5].
The illness is characterized by some following stages and their symptoms: For mild stage, illness is characterized by no sign of pneumonia and other serious illnesses like diarrhea and dyspnea. But the other symptoms include infection of an upper respiratory tract, fever, dry cough, sore throat, nasal discharge, sneezing, pain in muscles and headache sometimes [4,25]. In China, there were 81% of such cases were calculated.
In moderate illness, some patients showed symptoms of respiratory infections along with tachypnea, especially in children. There were no signs of pneumonia found in this type of patient.
In this serious illness, infection of respiratory along with dyspnea was found. High fever, hypoxia, and severe dyspnea were characterized by such patients. Some symptoms of cyanosis were also found in children.
While the critical condition of COVID-19 patients, failure of the respiratory trach, septic shock, and lung collapse is seen. Pulmonary edema and severe shortness of breath are also characterized. In this case organs, failure can also be seen. Reduce urination, high level of bilirubin, high lactate level, acidosis and thrombocytopenia were also observed [4,20,25].
Cardiovascular disease and COVID-19
Till now what we know, is that the mortality rate of this COVID-19 is very high in older individual’s i.e. older people underlying diseases, including Cardiovascular, respiratory and diabetic disease. However, some younger people’s death had been acknowledged after being hospitalized. Patients with long-term coronary artery disease, atherosclerotic disease, congenital heart disease, and cardiovascular diseases have a high level of risk of developing an acute infection as we have seen in MERS and Influenza [5,71]. Such acute coronary happening results in the severity and increase in myocardial demands activated by infections which cause injury to myocardial muscles and cause type 2 myocardial infarction and the circulating cytokines released in between the infection and stress conditioning could lead to plaque rupture and instability. Beside the COVID-19 infections, some other factor elevates the CVD (cardiovascular disorder) such as diabetes, immune function, dysregulated immunological status, hyperlipidemia impact [74,75].
Cardiovascular disease was a common disease and comorbidity in patients with COVID-19 predecessor in SARS and MERS. While in SARS, the existence of this CVD is 8%, and DM (Diabetes Mellitus) is 11% and both these comorbidity combine will increase the death risk twelve-fold [29,72,73,76]. In MERS, the DM and HTN (Hypertension) cases were almost 50% while it decreases so far in COD-19 to 30% [72]. The SARS-CoV has similar myocardial damage and pathogenicity which is caused by viruses surely increase the complexity and difficulty in understanding the patient and its treatment. Among the first 41 infected people with COVID-19, 5 are associated with myocardial injury, which demonstrated an increase in the quantity of level High sensitivity cardiac troponin I (hs-cTnI). Among 5, 4 people got admitted to ICU which demonstrates the intense myocardial injury nature. While the patient blood level is significantly higher in ICU patients compared to non-ICU patients.
There are two articles published in JAMA cardiology published by two different academic hospitals in Wuhan, China which is the starting point or epicenter of Coronavirus pandemic, provide the concept and understanding of the novel COVID-19 and its association with the myocardial injury [77, 78]. The 416 hospitalized patient COVID-19 positive confirmed, among them 82 [19.7%) showing myocardial injury have a highly elevated level of sensitivity of Troponin I (TnI). Myocardial injured patients have a higher mortality rate in hospitals [42 of 82 people [51.2%)) compared to those who do not mortality rate [15 of 335 [4.5%)) but among those myocardial injured patients, those who have higher TnI level have more mortality rate [78].
A very similar observation has been made by [78] in 187 confirmed COVID-19 hospitalized patients, along with 52 patients who had myocardial injury have a very high Troponin T (TnT) level. The mortality rate of this elevated TnT patient is 59.6% [31 of 52] compared to normal TnT level patients which are 8.9% [12 of 135]. Besides this observation, the patients who had cardiovascular disease with elevated TnT levels had a higher mortality rate in 25 of 36 [69.4%) in comparison to without prior cardiovascular disease, there is some elevation in TnT level in 6 of 16 [37.5%). The author provided an insight that observation of the TnT level associated with C-reactive protein level and N-terminal pro-B-type natriuretic peptide (NT-proBNP) which directly linked to ventricular dysfunction and inflammation severity. The available shows that there is a liberal increase in the NT-proBNP and TnT both levels in patients who are being hospitalized and followed a course to death whereas those with a favorable outcome with successful treatment, less severe illness and hospital discharge shows stable biomarker levels. [77] reported that patients which at the verge of myocardial injury risk as assessed by TnT or TnI level are mostly older and have higher coronary artery disease, hypertension, diabetes, and heart failure compared to those who had normal TnT or TnI level. Besides this hypertension and diabetes, the myocardial injured patients had more systematic inflammation i.e. higher C-reactive protein, procalcitonin and greater leukocyte counts and other biomarkers such as myoglobin, high creatine kinase and NT-proBNP. The patients confirmed with COVID-19 and myocardial injury show clear evidence of acute respiratory syndrome, the need for mechanical ventilators and required ICU treatment. This whole study gives us a clear image that confirmed COVID-19 patients with existed diabetics and myocardial injury have higher acuity of illness and develop a higher mortality rate in the short term [78].
In another research taken place in Wuhan, 138 patients confirmed with COVID-19, due to severe conditioning of 36 patients, were treated in ICU [14]. The patients in ICU have higher myocardial injury compared to those who are not treated in ICU (CK-MB level 18U/I vs 14U/I, TnI level 11.0 pg/ml vs 5.1 pg/ml), results suggest that those ICU patients have a higher myocardial injury. In confirmed COVID-19 patients, some patients went for cardiovascular symptoms of chest tightness and heart palpitation to the doctor, later on, found positive for COVID-19 [14]. Among the people who died of COVID-19, NHC (National Health Commission of China) stated that 11.8% of the patient without any prior CVD had a cardiac arrest, heart damage, and disorder of the immune system during hospitalization [14]. Therefore, in COVID-19 patients and the CVD symptoms frequency was high, due to immune response disorder and acute inflammatory response. The acute myocardial injury mechanism is might be due to ACE2, as it is widely present and expressed not only in the cardiovascular system also in the respiratory system, and it might have some heart injury role [79,80].
SARS-CoV-2 and ACE2
ACE-2 (Angiotensin-converting enzymes 2] is aminopeptidase membrane-bounded enzymes that have a very important role in immune and cardiovascular systems [75]. ACE2 involved in the development of hypertension, heart function, and diabetes mellitus. Besides that, ACE2 identified as the important receptor site for coronaviruses including SARS-CoV and SARS-CoV-2 [81-83]. The infection of COVID-19 activates ACE2 binding to the virus, which expressed majorly in lungs and hearts, so far that the COVID-19 majorly causes the respiratory infection and symptoms by invading the alveolar epithelium [81]. These symptoms of the respiratory infection get more severe with the patients having CVD, which might connect to the extra secretion of ACE2 compared to normal patients [84]. The ACE2 level artificially can be increased by the utilization of renin-angiotensin-aldosterone system inhibitors [84,85]. Given the information keep in mind about the ACE2 as it is a functional receptor for the COVID-19, antihypertension therapy should be observed with potential side effects and safety of patients [86,87]. Whether patients with COVID-19 and hypertension who are taking an ACE inhibitor or angiotensin-receptor blocker should switch to another antihypertensive drug [76,81].
The SARS-CoV-2 might infect the host by the ACE2 to cause infection of COVID-19 along causing myocardial damage, the thought mechanism of this infection is not clear. The patient with COVID-19 and CVD has an adverse prognosis, with a high risk of mortality and morbidity and the proper diagnosis and treatment should be done for these patients [76].
Pregnancy and COVID-19
At the end of 2019, the Novel coronavirus disease outbreak has become epidemic for china and now pandemic for the whole world, as per today 6th April, 1.275 Million has been infected with this novel virus among which 70,000 people died due to this virus (world meter site) [88]. During this outbreak, one of the major concerns related to pregnant women and neonatal babies. As the outbreak spread and unfolds, the control and prevention among pregnant women of COVID-19, and its vertical transmission risk have a major concern and issue [89]. More Evidence and study needed to develop and establish effective management and preventives and clinical strategies [90-92].
During Pregnancy, there is a change in physiological in their cardiopulmonary and immune system which makes pregnant women more likely to develop and host this virus and develop this severe infection [93,94]. Besides the image of this COVID-19 infection on women, there are some debates and concerns defining the potential effects on newborn and neonatal outcome and due to this factor, pregnant women need special attention to diagnosis, prevention, and management [94].
During influenza A H1N1 subtype outbreak, pregnant women hold only 1% while in the death toll, the number accounted for 5%. During the outbreak of SARS-CoV (severe acute respiratory syndrome coronavirus) in 2003, affecting more than 8000 persons in almost 26 countries with total death of 774, among which 12 pregnant women reported who develop this SARS-CoV. Among 12 pregnant women, 3 died during pregnancy, 4/7 have miscarriages, 4/5 pregnancies have preterm baby birth [95,96]. The pregnant women’s conditions infected with the SARS were worse from those who are not pregnant [89]. While for MERS which rose in 2012 starting from Saudi Arabia and spread to 27 countries infecting 11 pregnant women causing adverse clinical outcomes among them. The outcome of this virus on pregnant women includes premature delivery, maternal death, prenatal death, and intensive newborn baby care treatment [95]. But there is no confirmation about the vertical transmission of MERS from the infected mother to the baby. Both these viruses SARS and MERS responsible and caused severe complications during pregnancy and labor, including renal failure, miscarriages, prenatal death, endotracheal incubation while interesting the COVID-19 impact on pregnant women and newborn baby appear too small, and less severe [97].
So, for the analysis reveals that COVID-19 is unlike its predecessor strains among pregnant women, 38 pregnant women have positive results of COVID-19, no single lead to maternal death. And even with COVID-19, there were no cases of vertical transmission of SARS-CoV-2 from mothers to children. All neonatal specimens tested and the result was negative by rt-RCR. So, far this COVID-19 pandemic, there is no evidence and case of transplacental transmission during labor from mother to neonatal [96].
In Zhong nan Hospital Wuhan, 9 pregnant women with an age range from 26-40 with COVID-19 have been reviewed by medical staff and the women were tested by rt-PCR kits to perform double confirmation [98,99]. Most of these women are in 3rd trimester of pregnancy when they got infected with COVID-19. The symptoms they had was comprised of cough and fever only, with some other symptoms of diarrhea, throat, myalgia, malaise, and shortness of breath, with all these no chronic disorders, was caused in all pregnant women. The Chest scan and data of lab tests indicated that there are some numerous patchy shadows in the lung and there is an increase in the production of C-reactive protein. The complications of pregnancy that come after the onset of CoV infection include premature membrane rapture in 2/9 patients and in 2/9 fatal distress, but no patient died or developed pneumonia [98].
In Renmin Hospital Wuhan, sixteen pregnant women got infected with COVID-19, tested with dual fluorescence PCR and they got infected during the postpartum or mid-trimester period, shows similar findings except only of the women from eight other require ventilation and ICU for respiratory acute distress syndrome [96]. The results of these 16 pregnant women and their infants were compared to 45 pregnant women who do not have an infection with the age range from 24-36 years. The average weight of infants from Infected COVID-19 mother was 3.13 kg while for the non-infecting infants the average is 3.26 kg, no coherent difference between the parity, gravidity, gestational delivery age, intraoperative blood loss, and birthweight. While for the pregnant infected and non-infected to COVID-19 had no significant difference in gestational diabetes, premature membrane rupture, fetal distress, premature delivery, and other compress sutures. Among the mother with COVID-19, 10 infants have no infection of COVID-19, while 3 infants have bacterial pneumonia. These observations are also in line with what is being learned from COVID-19 pneumonia in pregnancy in other hospitals and these observation states that there is no vertical transfer of COVID-19 infection to the infant. This lack of transmission of maternal-fetal of COVID-19 is consistent with past outbreak experience of SARS and MERS happening in pregnant patients [96].
The Zheng [88]explained that the receptor angiotensin-converting enzyme 2 (ACE2] of COVID-19 shows the low level of expression in all types of cells of an early maternal-fetal interface which explains that there might know cell which is susceptible to COVID-19 in the maternal-fetal interface. This gives a clear indication that during pregnancy, it cannot lead to vertical transmission of transplacental. Though the data is limited at present most of the data available is from the third trimester, continue collecting data on COVID-19 pregnancy infection can enhance understanding of pregnancy disease and transplacental vertical transmission [88].
Management of COVID-19 in pregnancy
There are some general principle and ways to manage the pregnant women during COVID-19 which includes aggressive infections control procedure, isolation, SARS-CoV-2 testing, fluid overload avoidance, empiric antibiotics, coinfections, oxygen therapy if needed, early mechanic ventilation, monitoring of uterine and fatal contraction, doctorial team-based approach with specialty consultation delivery planning [24]. The team-based management is suggested for neonatal babies and pregnancies managed in the hospital facility and it should provide a determination to which to provide optimal care. The surveillance will be provided for the early detection of the worsening maternal illness and evidence obstetric complication are needed [24].
To observe the fatal maternal respiratory problem, heart rate patterns should be examined [88, 94]. From past experience during SARS and MERS, respiratory and heart failure might occur in pregnant women and the mechanical ventilator won’t support the normal flow of oxygen to the body. If in case, it happens to pregnant women having COVID-19 infection, literature provides the ECMO (extracorporeal membrane oxygenation) potential role in pregnancy [100].
So far, there is no antiviral medicine approved by the US Food and Drug Administration (FDA) to treat this infection of COVID-19, although the huge number of antivirals which was formulated for MERS in animals’ model can be evaluated for this COVID-19 virus, while the steroids like corticosteroids treatment should be avoided as there is no indication about these steroids for MERS-CoV clearance [101].
Management of neonatal born to mother having COVID-19
Due to limited data available about the COVID-19 infection and newborn babies and about its evaluations after the labor, but comparing with the SARS and MARS, so far there is no case that has been identified as the maternal to fetal or vertical transmission. Data from the recent case series published by [19,27,88]. About 45 of 18 women (19 infants) who were infected in the 3rd trimester with COVID-19, performed laboratory tests to verify infant with infection but no test was observed positive, so no vertical transmission [102]. Though some infants showed symptomatic (cyanosis in three, gastric bleeding in two, one died of multiple organ failure and shortness of breath in six).
So, the newborn baby expected to get infected either perinatally or in utero, so the newborn must be placed in self-isolation to avoid infection [93]. Breastmilk was tested for SARS-CoV-2 in six of the mothers reported by Chen, et al. [44] all specimens were negative [24]. So far there is no data available about the transmission of the infection through the breastmilk but in SARS there was a positive case as the virus was identified in breastmilk [93,103].
Diabetes mellitus and COVID-19
The COVID-19 virus directly causes respiratory infection and the diabetics patients are at risk of infections from other microbes and pathogens [104]. This site can be controlled through controlling the glycemic level, the good level of glycemic control will be benefited in controlling infections. Those patients who had diabetes must take vaccination of annual influenza and pneumococcal [105]. Besides this, the diabetic patients had a severe respiratory infection when infected with this COVID-19 virus. During the outbreak or pandemic of Influenza A 2009 (H1N1], MERS-CoV and SARS-CoV diabetes considered to be a significant factor for patients’ mortality [105,106].
So, far data obtained about the COVID-19 patients having diabetes at present is limited. In Wuhan, among 26 fatalities, 42.3% have diabetes, while in another study, 140 patients taken, but no considered threat was induced by COVID-19 to these diabetics and SARS-CoV-2 patients [107, 108]. An alternate study of 150 patients [82 recovered and 68 deaths) in Wuhan shows that the co-morbidities to be a significant predictor of mortality. The overall analysis of 11 different laboratory studies did not mention about the diabetics and raised glucose level as the predictor of mortality with COVID-19 infection [109,110]. Some hypotheses had been made by the scientists that those patients having the COVID-19 infection and treated with the ACE1-increasing doses, possess a higher risk for severe infection and even death, so it should be properly monitored. The data about this diabetic’s patients with COVID-19 is limited to make a proper response and theory [7,111,112].
Copper, a symbol of health
In China, the copper is called “qi” which means the symbol of health. In Egypt theology, the copper was said to be “ankh” the symbol for eternal life. Furthermore, the Phoenicians take this word as “Aphrodite — the goddess of love and beauty”.
The United States Environment Protection Agency has confirmed the antimicrobial effect of copper especially against the following agents; Enterobacter aerogenes, Escherichia coli O157: H7, Pseudomonas aeruginosa, Staphylococcus aureus, and Methicillin-resistant Staphylococcus aureus (MRSA). The doorknobs and push plates of brass in hospitals were found dirty and containing infectious agents but on the other hand knob doors and push plates of copper found cleaned [113].
The studies confirmed that when human coronavirus 229E brought in front of copper alloys, the virus becomes inactivated within a few minutes. Copper and zinc also found very effective against viruses. Exposure of viruses towards the copper completely destroys it. It changed the morphology of surface spike and other structural units. Copper should be used more in public places and where more people mostly gathered. This precautionary act will reduce the chance of virus transmission in public.
The research illustrated that when bacteria and some viruses are being exposed to the copper surface, they die. This is being proved in the lab as well as in the hospitals. It was noticed that the infection spread at a very low rate where there were cooper alloys components. So, learning from the data obtained, the hospital should be equipped with copper. This will cut down the treatment expenditures of a patient [74].
The cross-contamination is seemed to be reduced in a hospital when the pediatric patients were brought in contact with copper made surface in the ICU (intensive care unit). When it was compared with other metals which exhibited the less efficient. But, the relative risk reduction (RRR) was not found statistically significant [114].
In a study, the copper alloys proved very efficient in terms of antimicrobial activity in hospitals in an indoor environment. On the other hand, silver alloys exhibit low efficiency as compared to copper in a similar environment [115,116].
Preventions & precautions
The COVID-19 infection caused by the SARS-CoV-2 affects the lower respiratory tract with startling symptoms of flu, cough, muscle pain, tiredness, diarrhea, fever and then leads to major symptoms of acute respiration syndrome, septic shocks, pneumonia and sometimes leads to patient death. There is no specific vaccines or treatment for the treatment of this disease. There are some precautionary measures that need to be taken to prevent this infection to spread [117]. In the current scenario, the best strategies for the prevention of this COVID-19 virus are to take preventive measures and limit yourselves, because of the reason, the pandemic always increases if its R0 is greater than 1 (but COVID-19 have R0 value 2.2] [67]. To control this pandemic, it should be lesser than 1, so preventive measures are being taken to control this pandemic [118].
The preventive strategies basically engrossed on Patients’ isolation, self-isolation, social distancing, careful control of infection including the appropriate measures taken in diagnosis, and during clinical care. The WHO along with some organization provided us some recommendations to be followed to keep safe from this virus [119].
• Wash your hands frequently, often when you from outside or have contact with infected people.
• Maintain social distancing from people, especially when you are in an outer environment.
• Avoid any close contact with people who are suffering from a respiratory infection.
• Avoid any vulnerable contact with wild or farm animals.
• People who have respiratory or airway acute infection should keep a distance from them and the sneezing & coughing should be done on tissue paper, clothes, or any paper source and wash your hand as soon as possible.
• To protect yourself in the outer environment, always put masks to cover your mouth and hands to avoid contact with the virus.
• Young children, Immunocompromised, Old people, and pregnant women should avoid any public or private gathering to avoid the risk of infection.
• Strict hygiene measures must be carried out for control and prevention of infection in Hospitals, among medical staff.
• Avoid going out with unnecessary means or without any reasons.
• Quarantine yourself for almost 14 days to avoid any spread of infection.
This outbreak of COVID-19 rapidly swept across china and reached to more than 124 countries by 15 April 2020. Scientists and doctors working together made huge progress in the characterization of this virus and working broadly for the treatment and for developing the vaccination of this virus. In this study we have summarized different prospect of coronavirus as follow: Firstly, the transmission and symptomology of COVID-19 infection, explained that comparison to SARS and MERS coronavirus infection, COVID-19 has less virulence but more infectivity, originated from bat reservoir passing onto the unknown intermediate host, infect human via binding ACE2 receptor. Secondly, the infected population which is old or have medical condition i.e. History with fatal disease e.g. cardiovascular disorder, respiratory disorder, diabetes mellitus like requires more intensive care than normal patients. Thirdly, so far, there is no evidence found, that during childbirth or pregnancy it does not show any vertical transmission from mother to neonatal. Fourthly, the treatment which is done with antiviral drugs like Remdesivir, Ritonavir, and Chloroquine requires a more solid clinical trial and a solid statement is needed for their use. Besides all these, to minimize the risk of COVID-19 precautions need to be taken.
- Organization WH. WHO Director-General’s remarks at the media briefing on 2019-nCoV on 11 February 2020. Internet] World Health Organization. 2020.
- Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. The Lancet. 2020; 395: 470-473. PubMed: https://pubmed.ncbi.nlm.nih.gov/31986257/
- Sohrabi C, Alsafi Z, O’Neill N, Khan M, Kerwan A, Al-Jabir A, et al. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int J Surg. 2020. PubMed: https://pubmed.ncbi.nlm.nih.gov/32112977/
- Kogan A, Segel MJ, Ram E, Raanani E, Peled-Potashnik Y, et al. Acute Respiratory Distress Syndrome following Cardiac Surgery: Comparison of the American-European Consensus Conference Definition versus the Berlin Definition. Respiration. 2019; 97: 518-524. PubMed: https://pubmed.ncbi.nlm.nih.gov/30650409/
- Huang C, Wang Y, Li X, Ren L, Zhao J, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020; 395: 497-506. PubMed: https://pubmed.ncbi.nlm.nih.gov/31986264/
- Hui DS, Azhar EI, Madani TA, Ntoumi F, Kock R, et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020; 91: 264. PubMed: https://pubmed.ncbi.nlm.nih.gov/31953166/
- Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020. PubMed: https://pubmed.ncbi.nlm.nih.gov/32091533/
- Coronavirus disease 2019 (COVID-19) Situation Report – 86.
- Bai Y, Yao L, Wei T, Tian F, Jin DY, et al. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020; 323: 1406-1407. PubMed: https://pubmed.ncbi.nlm.nih.gov/32083643/
- Kahn JS, McIntosh K. History and recent advances in coronavirus discovery. Pediatr Infect Dis J. 2005; 24: S223-S227. PubMed: https://pubmed.ncbi.nlm.nih.gov/16378050/
- Savarino A, Boelaert JR, Cassone A, Majori G, Cauda R. Effects of chloroquine on viral infections: an old drug against today's diseases. Lancet Infect Dis. 2003; 3: 722-727. PubMed: https://pubmed.ncbi.nlm.nih.gov/14592603/
- Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Advan Res. 2020. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7113610/
- Tyrrell D, Bynoe M. Cultivation of viruses from a high proportion of patients with colds. Lancet. 1966: 76-77. PubMed: https://pubmed.ncbi.nlm.nih.gov/4158999/
- Wang D, Hu B, Hu C, Zhu F, Liu X, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020; 323: 1061-1069. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/pmc7042881/
- Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005; 79: 884-895. PubMed: https://pubmed.ncbi.nlm.nih.gov/15613317/
- Woo PC, Lau SK, Tsoi HW, Huang Y, Poon RW, et al. Clinical and molecular epidemiological features of coronavirus HKU1–associated community-acquired pneumonia. J Infect Dis. 2005; 192: 1898-1907. PubMed: https://pubmed.ncbi.nlm.nih.gov/16267760
- Woo PC, Wang M, Lau SK, Xu H, Poon RW, Guo R, et al. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J Virol. 2007; 81: 1574-1585. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2612373/
- Bosis S, Esposito S, Niesters HG, Tremolati E, Pas S, et al. Coronavirus HKU1 in an Italian pre-term infant with bronchiolitis. J Clin Virol. 2007; 38: 251-253. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/17320791
- Han CL, Yang F, Zhang TJ. 2001.
- Chiu SS, Hung Chan K, Wing Chu K, Kwan SW, et al. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin Infect Dis. 2005; 40: 1721-1729. PubMed: https://pubmed.ncbi.nlm.nih.gov/15909257/
- Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Coronaviruses: Springer; 2015; 1-23. PubMed: https://pubmed.ncbi.nlm.nih.gov/25720466/
- Colson P, Rolain J-M, Lagier J-C, Brouqui P, Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int J Antimicrob Agents. 2020; 105932. PubMed: https://pubmed.ncbi.nlm.nih.gov/32145363/
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, et al. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. Nature. 2020; 579: 270-273. PubMed: https://pubmed.ncbi.nlm.nih.gov/32015507/
- Paules CI, Marston HD, Fauci AS. Coronavirus infections—more than just the common cold. JAMA. 2020; 323: 707-708. PubMed: https://pubmed.ncbi.nlm.nih.gov/31971553/
- Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A. 2005; 102: 14040-14045. PubMed: https://pubmed.ncbi.nlm.nih.gov/16169905/
- Hu B, Ge X, Wang LF, Shi Z. Bat origin of human coronaviruses. Virol J. 2015; 12: 221.
- Zhu N, Zhang D, Wang W, Li X, Yang B, et al. China Novel Coronavirus Investigating and Research Team. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020; 382: 727-733. PubMed: https://pubmed.ncbi.nlm.nih.gov/31978945/
- Cheever FS, Daniels JB, Pappenheimer AM, Bailey OT. A murine virus (JHM) causing disseminated encephalomyelitis with extensive destruction of myelin: I. Isolation and biological properties of the virus. J Experimen Med. 1949; 90: 181-210. PubMed: https://pubmed.ncbi.nlm.nih.gov/18137294
- Guan Y, Zheng B, He Y, Liu X, Zhuang Z, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003; 302: 276-268. PubMed: https://pubmed.ncbi.nlm.nih.gov/12958366/
- Hamre D, Procknow JJ. A new virus isolated from the human respiratory tract. Proc Society Experimen Biol Med. 1966; 121: 190-193. PubMed: https://pubmed.ncbi.nlm.nih.gov/4285768/
- Doyle I. LP and Hutchings, LM A transmissible gastroenteritis in pigs. J Am Vet Med Assoc. 1946; 108: 257-259. PubMed: https://pubmed.ncbi.nlm.nih.gov/21020443/
- Chu D, Peiris J, Chen H, Guan Y, Poon LL. Genomic characterizations of bat coronaviruses (1A, 1B and HKU8) and evidence for co-infections in Miniopterus bats. J General Virol. 2008; 89: 1282-1287. PubMed: https://pubmed.ncbi.nlm.nih.gov/18420807/
- Esper F, Weibel C, Ferguson D, Landry ML, Kahn JS. Coronavirus HKU1 infection in the United States. Emerging Infect Dis. 2006; 12: 775. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3374449/
- Gerna G, Percivalle E, Sarasini A, Campanini G, Piralla A, Rovida F, et al. Human respiratory coronavirus HKU1 versus other coronavirus infections in Italian hospitalised patients. J Clini Virol. 2007; 38: 244-250. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3185465/
- Kupfer B, Simon A, Jonassen C, Viazov S, Ditt V, et al. Two cases of severe obstructive pneumonia associated with an HKU1-like coronavirus. Euro J Med Res. 2007; 12: 134.
- Lau SK, Poon RW, Wong BH, Wang M, Huang Y, et al. Coexistence of different genotypes in the same bat and serological characterization of Rousettus bat coronavirus HKU9 belonging to a novel Betacoronavirus subgroup. J Virol. 2010; 84: 11385-11394.
- Lau SK, Woo PC, Yip CC, Tse H, Tsoi HW, et al. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J Clin Microbiol. 2006; 44: 2063-20671. PubMed: https://pubmed.ncbi.nlm.nih.gov/16757599/
- Vabret A, Dina J, Gouarin S, Petitjean J, Corbet S, Freymuth F. Detection of the new human coronavirus HKU1: a report of 6 cases. Clin Infect Dis. 2006; 42: 634-639. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7107802/
- Sloots TP, McErlean P, Speicher DJ, Arden KE, Nissen MD, et al. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clini Virol. 2006; 35: 99-102. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/16257260
- Woo PC, Lau SK, Yip CC, Huang Y, Tsoi HW, et al. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J Virol. 2006; 80: 7136-7145. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1489027/
- Lau SK, Woo PC, Li KS, Huang Y, Wang M, et al. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. 2007; 367: 428-439. PubMed: https://pubmed.ncbi.nlm.nih.gov/17617433/
- Arden KE, Nissen MD, Sloots TP, Mackay IM. New human coronavirus, HCoV‐NL63, associated with severe lower respiratory tract disease in Australia. J Med Virol. 2005; 75: 455-462. PubMed: https://pubmed.ncbi.nlm.nih.gov/15648064/
- Mihindukulasuriya KA, Wu G, Leger JS, Nordhausen RW, Wang D. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J Virol. 2008; 82: 5084-5098. PubMed: https://pubmed.ncbi.nlm.nih.gov/18353961/
- Beaudette F. Cultivation of the virus of infectious bronchitis. J Am Vet Med Assoc. 1937; 90: 51-60.
- Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019; 17: 181-192. PubMed: https://pubmed.ncbi.nlm.nih.gov/30531947/
- van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, et al. Identification of a new human coronavirus. Nat Med. 2004; 10: 368-373. PubMed: https://pubmed.ncbi.nlm.nih.gov/15034574/
- Wu A, Peng Y, Huang B, Ding X, Wang X, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020. PubMed: https://pubmed.ncbi.nlm.nih.gov/32035028/
- Li B, Si HR, Zhu Y, Yang XL, Anderson DE, et al. Discovery of bat coronaviruses through surveillance and probe capture-based next-generation sequencing. Msphere. 2020; 5.
- Gralinski L, Menachery V. Return of the coronavirus: 2019-nCoV. Viruses. J Popul Ther Clin Pharmacol. 2020; 27: e19-e22. PubMed: https://pubmed.ncbi.nlm.nih.gov/31991541/
- Fouchier RA, Hartwig NG, Bestebroer TM, Niemeyer B, de Jong JC, et al. A previously undescribed coronavirus associated with respiratory disease in humans. Proceedings of the National Academy of Sciences. 2004; 101: 6212-6216. PubMed: https://pubmed.ncbi.nlm.nih.gov/15073334/
- Tang X, Zhang J, Zhang S, Wang P, Fan X, et al. Prevalence and genetic diversity of coronaviruses in bats from China. J Virol. 2006; 80: 7481-7490. PubMed: https://pubmed.ncbi.nlm.nih.gov/16840328/
- Jendrach M, Thiel V, Siddell S. Characterization of an internal ribosome entry site within mRNA 5 of murine hepatitis virus. Archives of virology. 1999; 144: 921-933. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/10416375
- Esposito S, Bosis S, Niesters HG, Tremolati E, Begliatti E, et al. Impact of human coronavirus infections in otherwise healthy children who attended an emergency department. J Med Virol. 2006; 78: 1609-1615. PubMed: https://pubmed.ncbi.nlm.nih.gov/17063525/
- Ebihara T, Endo R, Ma X, Ishiguro N, Kikuta H. Detection of human coronavirus NL63 in young children with bronchiolitis. J Med Virol. 2005; 75: 463-465. PubMed: https://pubmed.ncbi.nlm.nih.gov/15648061/
- Koetz A, Nilsson P, Lindén MV, Van Der Hoek L, Ripa T. Detection of human coronavirus NL63, human metapneumovirus and respiratory syncytial virus in children with respiratory tract infections in south-west Sweden. Clin Microbiol Infect. 2006; 12: 1089-1096. PubMed: https://pubmed.ncbi.nlm.nih.gov/17002608/
- Suzuki A, Okamoto M, Ohmi A, Watanabe O, Miyabayashi S, Nishimura H. Detection of human coronavirus-NL63 in children in Japan. Pediat Infect Dis J. 2005; 24: 645-646. PubMed: https://pubmed.ncbi.nlm.nih.gov/15999010/
- Xu X, Chen P, Wang J, Feng J, Zhou H, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Science China Life Sciences. 2020; 63: 457-460. PubMed: https://pubmed.ncbi.nlm.nih.gov/32009228/
- Organization WH. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance, 2 March 2020. World Health Organization. 2020.
- Corman VM, Ithete NL, Richards LR, Schoeman MC, Preiser W, et al. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J Virol. 2014; 88: 11297-11303. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4178802/
- Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. The Lancet. 2020; 395: 507-513. PubMed: https://pubmed.ncbi.nlm.nih.gov/32007143/
- Coronavirus and pets: faqs for owners.
- USDA Statement on the Confirmation of COVID-19 in a Tiger in New York.
- Acconcia A, D’Amato M, Martina R. Tax evasion and corruption in tax administration. J Public Economics. 2003; 105: 1-2.
- Tabriz AG, Hermida MA, Leslie NR, Shu W. Three-dimensional bioprinting of complex cell laden alginate hydrogel structures. Biofabrication. 2015; 7: 045012. PubMed: https://pubmed.ncbi.nlm.nih.gov/26689257/
- Li Q, Guan X, Wu P, Wang X, Zhou L, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Eng J Med. 2020. PubMed: https://pubmed.ncbi.nlm.nih.gov/31995857/
- Wu F, Zhao S, Yu B. A new coronavirus associated with human respiratory disease in China. Nature. 2020; 579: 265–269.. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7094943/
- Chan JW, Kok KH, Zhu Z, Chu H, To KKW, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging Microbes Infect. 2020; 9: 221-236. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7067204/
- Liu Z, Xiao X, Wei X, Li J, Yang J, et al. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS‐CoV‐2. J Med Virol. 2020; 92: 595-601. PubMed: https://pubmed.ncbi.nlm.nih.gov/32100877/
- Paraskevis D, Kostaki EG, Magiorkinis G, Panayiotakopoulos G, Sourvinos G, et al. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infection, Genetics and Evolution. 2020; 79: 104212. PubMed: https://pubmed.ncbi.nlm.nih.gov/32004758/
- Kang CK, Song KH, Choe PG, Park WB, Bang JH, et al. Clinical and epidemiologic characteristics of spreaders of Middle East respiratory syndrome coronavirus during the 2015 outbreak in Korea. J Korean Med Sci. 2017; 32: 744-749. PubMed: https://pubmed.ncbi.nlm.nih.gov/28378546/
- Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, et al. Clinical characteristics of coronavirus disease 2019 in China. N Eng J Med. 2020; 382: 1708-1720. PubMed: https://pubmed.ncbi.nlm.nih.gov/32109013/
- Chowell G, Abdirizak F, Lee S, Lee J, Jung E, et al. Transmission characteristics of MERS and SARS in the healthcare setting: a comparative study. BMC Med. 2015; 13: 210. PubMed: https://pubmed.ncbi.nlm.nih.gov/26336062/
- Bauch CT, Lloyd-Smith JO, Coffee MP, Galvani AP. Dynamically modeling SARS and other newly emerging respiratory illnesses: past, present, and future. Epidemiology. 2005: 791-801. PubMed: https://pubmed.ncbi.nlm.nih.gov/16222170/
- Daskalopoulou SS, Rabi DM, Zarnke KB, Dasgupta K, Nerenberg K, et al. The 2015 Canadian Hypertension Education Program recommendations for blood pressure measurement, diagnosis, assessment of risk, prevention, and treatment of hypertension. Canadian J Cardiol. 2015; 31: 549-568. PubMed: https://pubmed.ncbi.nlm.nih.gov/25936483
- Zheng YY, Ma YT, Zhang JY, Xie X. COVID-19 and the cardiovascular system. Nat Rev Cardiol. 2020; 17: 259-260. PubMed: https://pubmed.ncbi.nlm.nih.gov/32139904/
- Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat Med. 2005; 11: 875-879. PubMed: https://pubmed.ncbi.nlm.nih.gov/16007097/
- Shi S, Qin M, Shen B, Cai Y, Liu T, et al. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol. 2020; 25; 5: 802-810. PubMed: https://pubmed.ncbi.nlm.nih.gov/32211816/
- Guo T, Fan Y, Chen M, Wu X, Zhang L, et al. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19). JAMA Cardiol. 2020; 5: 1-8. PubMed: https://pubmed.ncbi.nlm.nih.gov/32219356
- Lillie PJ, Samson A, Li A, Adams K, Capstick R, Barlow GD, et al. Novel coronavirus disease (Covid-19): the first two patients in the UK with person to person transmission. J Infect. 2020; 80: 578-606 PubMed: https://pubmed.ncbi.nlm.nih.gov/32119884/
- Alhogbani T. Acute myocarditis associated with novel Middle East respiratory syndrome coronavirus. Ann Saudi Med. 2016; 36: 78-80. PubMed: https://pubmed.ncbi.nlm.nih.gov/26922692/
- Imai Y, Kuba K, Rao S, Huan Y, Guo F, et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005; 436: 112-116. PubMed: https://pubmed.ncbi.nlm.nih.gov/16001071/
- Gurwitz D. Angiotensin receptor blockers as tentative SARS‐CoV‐2 therapeutics. Drug Dev Res. 2020. PubMed: https://pubmed.ncbi.nlm.nih.gov/32129518/
- Ishiyama Y, Gallagher PE, Averill DB, Tallant EA, Brosnihan KB, et al. Upregulation of angiotensin-converting enzyme 2 after myocardial infarction by blockade of angiotensin II receptors. Hypertension. 2004; 43: 970-976. PubMed: https://pubmed.ncbi.nlm.nih.gov/15007027/
- Ferrario CM, Jessup J, Chappell MC, Averill DB, Brosnihan KB, et al. Effect of angiotensin-converting enzyme inhibition and angiotensin II receptor blockers on cardiac angiotensin-converting enzyme 2. Circulation. 2005; 111: 2605-2610. PubMed: https://pubmed.ncbi.nlm.nih.gov/15897343/
- Ocaranza MP, Palomera C, Román M, Bargetto J, Lavandero S, et al. Effect of hypertension on angiotensin-(1–7) levels in rats with different angiotensin-I converting enzyme polymorphism. Life Sciences. 2006; 78: 1535-1542.
- Walters TE, Kalman JM, Patel SK, Mearns M, Velkoska E, et al. Angiotensin converting enzyme 2 activity and human atrial fibrillation: increased plasma angiotensin converting enzyme 2 activity is associated with atrial fibrillation and more advanced left atrial structural remodelling. Ep Europace. 2017; 19: 1280-1287. PubMed: https://pubmed.ncbi.nlm.nih.gov/27738071/
- Burchill LJ, Velkoska E, Dean RG, Griggs K, Patel SK, et al. Combination renin–angiotensin system blockade and angiotensin-converting enzyme 2 in experimental myocardial infarction: implications for future therapeutic directions. Clin Sci. 2012; 123: 649-658. PubMed:
- Zhu H, Wang L, Fang C, Peng S, Zhang L, et al. Clinical analysis of 10 neonates born to mothers with 2019-nCoV pneumonia. Translational Pediatrics. 2020; 9: 51-60. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7036645/
- Alvarado M, Schwartz D. Zika virus in pregnancy, microcephaly and maternal and fetal health: What we think, what we know, and what we think we know. Arch Pathol Lab Med. 2016; 141: 26-32. PubMed: https://pubmed.ncbi.nlm.nih.gov/27636525/
- Zhu N, Zhang D, Wang W, Li X, Yang B, et al. China Novel Coronavirus Investigating and Research Team. N Engl J Med. 2020; 382: 727-733. PubMed: https://pubmed.ncbi.nlm.nih.gov/31978945/
- Huang C. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with. 2019: 497-506. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7159299/
- Michalowski W, Rubin S, Slowinski R, Wilk S. Mobile clinical support system for pediatric emergencies. Decision Support Systems. 2003; 36: 161-176.
- Rasmussen SA, Kissin DM, Yeung LF, MacFarlane K, Chu SY, et al. Preparing for influenza after 2009 H1N1: special considerations for pregnant women and newborns. Am J Obstet Gynecol. 2011; 204: S13-S20. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6771262/
- Rasmussen SA, Smulian JC, Lednicky JA, Wen TS, Jamieson DJ. Coronavirus Disease 2019 (COVID-19) and Pregnancy: What obstetricians need to know. Am J Obstet Gynecol. 2020; 222: 415-426. PubMed: https://pubmed.ncbi.nlm.nih.gov/32105680/
- Schwartz DA, Graham AL. Potential maternal and infant outcomes from (Wuhan) coronavirus 2019-nCoV infecting pregnant women: lessons from SARS, MERS, and other human coronavirus infections. Viruses. 2020; 12: 194. PubMed: https://pubmed.ncbi.nlm.nih.gov/32050635/
- Schwartz DA. An analysis of 38 pregnant women with COVID-19, their newborn infants, and maternal-fetal transmission of SARS-CoV-2: maternal coronavirus infections and pregnancy outcomes. Arch Pathol Lab Med. 2020. PubMed: https://pubmed.ncbi.nlm.nih.gov/32180426/
- Maxwell C, McGeer A, Tai KFY, Sermer M. No. 225-Management Guidelines for Obstetric Patients and Neonates Born to Mothers With Suspected or Probable Severe Acute Respiratory Syndrome (SARS). J Obstet Gynaecol Canada. 2017; 39: e130-e137. PubMed: https://pubmed.ncbi.nlm.nih.gov/28729104/
- Wang X, Zhou Z, Zhang J, Zhu F, Tang Y, et al. A case of 2019 Novel Coronavirus in a pregnant woman with preterm delivery. Clin Infect Dis. 2020; ciaa200 PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7108126/
- Chen H, Guo J, Wang C, Luo F, Yu X, et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. The Lancet. 2020; 395: 809-815. PubMed: https://pubmed.ncbi.nlm.nih.gov/32151335/
- Pacheco LD, Saade GR, Hankins GD, editors. Extracorporeal membrane oxygenation (ECMO) during pregnancy and postpartum. Semin Perinatol. 2018: 42: 21-25. PubMed: https://pubmed.ncbi.nlm.nih.gov/29179956/
- Arabi YM, Mandourah Y, Al-Hameed F, Sindi AA, Almekhlafi GA, et al. Corticosteroid therapy for critically ill patients with Middle East respiratory syndrome. Am J Respirat Critical Care Med. 2018; 197: 757-767. PubMed: https://pubmed.ncbi.nlm.nih.gov/29161116/
- D’Amore R. Can coronavirus pass from mother to baby? Maybe, but experts need more research. Global News. 2020; 2020.
- Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020; 94: e00127-120. PubMed: https://pubmed.ncbi.nlm.nih.gov/31996437/
- Lane HC, Marston HD, Fauci AS. Conducting clinical trials in outbreak settings: points to consider. Clinical trials. 2016; 13: 92-95. PubMed: https://www.ncbi.nlm.nih.gov/books/NBK441674/
- Yang J, Feng Y, Yuan M, Yuan S, Fu H, et al. Plasma glucose levels and diabetes are independent predictors for mortality and morbidity in patients with SARS. Diabetic Med. 2006; 23: 623-628. PubMed: https://pubmed.ncbi.nlm.nih.gov/16759303/
- Alene KA, Viney K, Gray DJ, McBryde ES, Wagnew M, et al. Mapping tuberculosis treatment outcomes in Ethiopia. BMC Infect Dis. 2019; 19: 474. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/31138129
- Deng SQ, Peng HJ. Characteristics of and public health responses to the coronavirus disease 2019 outbreak in China. J Clin Med. 2020; 9: 575. PubMed: https://pubmed.ncbi.nlm.nih.gov/32093211/
- Zhang JJ, Dong X, Cao YY, Yuan YD, Yang YB, et al. Clinical characteristics of 140 patients infected with SARS‐CoV‐2 in Wuhan, China. Allergy. 2020; 75: 1730-1741 PubMed: https://pubmed.ncbi.nlm.nih.gov/32077115/
- Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020: 46: 846-848. PubMed: https://pubmed.ncbi.nlm.nih.gov/32125452/
- Lippi G, Plebani M. Laboratory abnormalities in patients with COVID-2019 infection. Clin Chem Lab Med. 2020; 58: 1131-1134. PubMed: https://pubmed.ncbi.nlm.nih.gov/32119647/
- Sun M, Yang J, Sun Y, Su G. Inhibitors of RAS might be a good choice for the therapy of COVID-19 pneumonia. Zhonghua Jie He He Hu Xi Za Zhi. 2020; 43: E014-014E. PubMed: https://pubmed.ncbi.nlm.nih.gov/32061198/
- Phadke M, Saunik S. Use of angiotensin receptor blockers such as Telmisartan, Losartsan in nCoV Wuhan Corona Virus infections–Novel mode of treatment. BMJ. 2020; 368: 406.
- Lamanna C, Mallette MF. Basic Bacteriology: Its Biological and Chemical Background. Basic Bacteriology: Its Biological and Chemical Background. 1965; 3.
- Schmidt MG, von Dessauer B, Benavente C, Benadof D, Cifuentes P, et al. Copper surfaces are associated with significantly lower concentrations of bacteria on selected surfaces within a pediatric intensive care unit. Am J Infect Control. 2016; 44: 203-209. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/26553403
- Henderson E, Michels J, Sagel J. Flexible film plate-mat bag. Google Patents; 2009.
- Michels H, Noyce J, Keevil CW. Effects of temperature and humidity on the efficacy of methicillin‐resistant Staphylococcus aureus challenged antimicrobial materials containing silver and copper. Lett Appl Microbiol. 2009; 49: 191-195. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2779462/
- Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY. Coronaviruses—drug discovery and therapeutic options. Nature reviews Drug discovery. 2016; 15: 327-347. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7097181/
- Thompson R. Pandemic potential of 2019-nCoV. The Lancet Infect Dis. 2020; 20: 280. PubMed: https://pubmed.ncbi.nlm.nih.gov/32043978/
- Coronavirus disease (COVID-19) advice for the public: When and how to use masks.